Molecular Detection of Antibiotic Resistance Genes Using Respiratory Sample from Pneumonia Patients
- PMID: 40426568
- PMCID: PMC12108368
- DOI: 10.3390/antibiotics14050502
Molecular Detection of Antibiotic Resistance Genes Using Respiratory Sample from Pneumonia Patients
Abstract
Introduction/objectives: Antibiotic resistance makes the treatment of pneumonia challenging. Effective management depends on accurate diagnostic techniques to identify resistance genes and customize drugs. This study primarily aimed to identify antibiotic resistance genes in respiratory samples from patients with pneumonia using polymerase chain reaction (PCR) to determine the prevalence of specific resistance genes and analyze clinical factors contributing to antibiotic resistance, as well as to provide actionable insights into resistance patterns in Jordan and support efforts to improve pneumonia management.
Methods: This retrospective observational study included 114 patients who were diagnosed with pneumonia. Clinical data, including prior antibiotic exposure and treatment history, were collected. PCR diagnostics were used to detect resistance genes in respiratory samples. In this study, we evaluated 14 antibiotic resistance genes in pneumonia pathogens, highlighting their diverse resistance mechanisms.
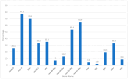
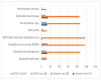
Results: Mec A was the most frequently detected gene, appearing in 87 samples (77.3%). Additionally, Tem in 80 samples (70.2%), Oxa-48-like in 15 samples (13.2%), and Ctx-M-1 in 38 samples (33.3%) were among the most commonly detected genes. In contrast, Oxa-40-like (7.0%), Vim (8.8%), and Imp (4.4%) genes exhibited a lower prevalence. The Oxa-51-like gene showed the only significant association with ertapenem resistance (p-value = 0.046). Further analysis revealed statistically significant associations between Mec A and methicillin resistance (p < 0.001), underscoring its critical role. However, other genes, such as Oxa-40-like and Oxa-48-like, showed no significant correlation with the antibiotic resistance patterns of imipenem and meropenem (p > 0.05).
Conclusions: This study demonstrates the utility of PCR-based diagnostics for detecting resistance genes and highlights the critical clinical factors associated with antibiotic resistance in patients with pneumonia. These findings underscore the importance of integrating molecular diagnostics into routine care to improve treatment outcomes and combat the growing threat of antibiotic resistance in Jordan. This highlights PCR's value in guiding effective treatment strategies and addressing multidrug-resistant pneumonia.
Keywords: antibiotic-resistant genes; antimicrobial; epidemiology; hospital; pneumonia.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures
Similar articles
-
Molecular detection of OXA-48 and NDM-1 carbapenemase genes among clinical isolates of Klebsiella pneumoniae recovered from patients attending a private tertiary hospital in Southwestern Nigeria.BMC Infect Dis. 2024 Sep 13;24(1):970. doi: 10.1186/s12879-024-09869-x. BMC Infect Dis. 2024. PMID: 39271986 Free PMC article.
-
Molecular Detection of Carbapenemase-Encoding Genes in Multidrug-Resistant Acinetobacter baumannii Clinical Isolates in South Africa.Int J Microbiol. 2020 Jun 13;2020:7380740. doi: 10.1155/2020/7380740. eCollection 2020. Int J Microbiol. 2020. PMID: 32612659 Free PMC article.
-
High prevalence of oxacillinases in clinical multidrug-resistant Acinetobacter baumannii isolates from the Tshwane region, South Africa - an update.BMC Infect Dis. 2015 Nov 14;15:521. doi: 10.1186/s12879-015-1246-8. BMC Infect Dis. 2015. PMID: 26573617 Free PMC article.
-
Characterization of Oxacillinase and Metallo-β-Lactamas Genes and Molecular Typing of Clinical Isolates of Acinetobacter baumannii in Ahvaz, South-West of Iran.Jundishapur J Microbiol. 2016 Feb 13;9(5):e32388. doi: 10.5812/jjm.32388. eCollection 2016 May. Jundishapur J Microbiol. 2016. PMID: 27540456 Free PMC article.
-
Sulopenem: An Intravenous and Oral Penem for the Treatment of Urinary Tract Infections Due to Multidrug-Resistant Bacteria.Drugs. 2022 Apr;82(5):533-557. doi: 10.1007/s40265-022-01688-1. Epub 2022 Mar 16. Drugs. 2022. PMID: 35294769 Review.
References
LinkOut - more resources
Full Text Sources
Miscellaneous