Multi-Omics-Based Analysis of the Effect of Longevity Genes on the Immune Relevance of Colorectal Cancer
- PMID: 40426913
- PMCID: PMC12109330
- DOI: 10.3390/biomedicines13051085
Multi-Omics-Based Analysis of the Effect of Longevity Genes on the Immune Relevance of Colorectal Cancer
Abstract
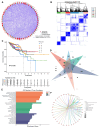
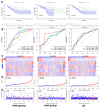
Background: Colorectal cancer (CRC) ranks as the third most prevalent cancer globally, with its incidence and recurrence rates steadily rising. To explore the relationship between CRC and longevity-associated genes (LAGs), and to offer new therapeutic avenues for CRC treatment, we developed a prognostic model based on these genes to predict the outcomes for CRC patients. Additionally, we conducted an immune correlation analysis. Methods: We conducted a comprehensive analysis of the effects of 81 LAGs in CRC by integrating multiple omics datasets. This analysis led to the identification of two distinct molecular subtypes and revealed that alterations in LAGs across various layers were linked to clinicopathological features, prognosis, and cell infiltration characteristics within the tumor microenvironment (TME). The training and validation cohorts for the models were derived from the TCGA-COAD, TCGA-READ, and GSE35279 datasets. Subsequently, we developed a risk score model, and the Kaplan-Meier method was employed to estimate overall survival (OS). Ultimately, we established a prognostic model based on five LAGs: BEDN3, EXOC3L2, CDKN2A, IL-13, and CAPN9. Furthermore, we assessed the correlations between the risk score and factors such as immune cell infiltration, microsatellite instability, and the stem cell index. Results: Our comprehensive bioinformatics analysis revealed a strong association between longevity genes and CRC. The risk score derived from the five newly identified LAGs was determined to be an independent prognostic factor for CRC. Patients categorized by this risk score demonstrated significant differences in immune status and microsatellite instability. Conclusions: Our comprehensive multi-omic analysis of LAGs highlighted their potential roles in the tumor immune microenvironment, clinicopathological features, and prognosis, offering new insights for the treatment of CRC.
Keywords: bioinformatics; colorectal cancer; immunization; longevity; multi-omics.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures




Similar articles
-
Comprehensive analysis of cuproptosis-related genes involved in prognosis and tumor microenvironment infiltration of colorectal cancer.Transl Cancer Res. 2024 Sep 30;13(9):4555-4573. doi: 10.21037/tcr-24-546. Epub 2024 Sep 27. Transl Cancer Res. 2024. PMID: 39430854 Free PMC article.
-
Identification of prognostic immune-related gene signature associated with tumor microenvironment of colorectal cancer.BMC Cancer. 2021 Aug 8;21(1):905. doi: 10.1186/s12885-021-08629-3. BMC Cancer. 2021. PMID: 34364366 Free PMC article.
-
Single-cell and Multi-omics Analysis Confirmed the Signature and Potential Targets of Cuproptosis in Colorectal Cancer.J Cancer. 2025 Jan 13;16(4):1264-1280. doi: 10.7150/jca.104702. eCollection 2025. J Cancer. 2025. PMID: 39895793 Free PMC article.
-
Role of ferroptosis-related genes in prognostic prediction and tumor immune microenvironment in colorectal carcinoma.PeerJ. 2021 Jul 14;9:e11745. doi: 10.7717/peerj.11745. eCollection 2021. PeerJ. 2021. PMID: 34316400 Free PMC article.
-
Mapk14 is a Prognostic Biomarker and Correlates with the Clinicopathological Features and Immune Infiltration of Colorectal Cancer.Front Cell Dev Biol. 2022 Jan 24;10:817800. doi: 10.3389/fcell.2022.817800. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 35141222 Free PMC article.
References
-
- Wang H., Gou R., Chen J., Wang Q., Li X., Chang J., Chen H., Wang X., Wan G. Catalase-positive Staphylococcus epidermidis based cryo-millineedle platform facilitates the photo-immunotherapy against colorectal cancer via hypoxia improvement. J. Colloid Interface Sci. 2024;676:506–520. doi: 10.1016/j.jcis.2024.07.145. - DOI - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

