PARP Inhibitors in Ovarian Cancer: Resistance Mechanisms, Clinical Evidence, and Evolving Strategies
- PMID: 40426953
- PMCID: PMC12108591
- DOI: 10.3390/biomedicines13051126
PARP Inhibitors in Ovarian Cancer: Resistance Mechanisms, Clinical Evidence, and Evolving Strategies
Abstract
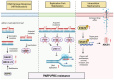
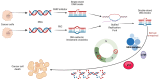
The introduction of poly (ADP-ribose) polymerase inhibitors (PARPi) into the management of ovarian cancer has transformed the treatment landscape for patients affected by this malignancy. However, as the use of PARPi expands into both frontline maintenance and recurrence settings, the emergence of drug resistance has become a significant clinical challenge in the treatment of these patients. Although platinum-based chemotherapy (PBC) and PARPi act through different mechanisms-PBC causes DNA damage while PARPi blocks its repair-both depend on the integrity of DNA damage repair (DDR) pathways, leading to overlapping mechanisms of resistance. Here, we review the key resistance mechanisms shared by PARPi and PBC, and then we discuss their clinical implications in the management of patients with ovarian cancer. We also examine clinical rationale supporting the hypothesis that prior PARPi exposure may reduce the efficacy of subsequent PBC in patients experiencing a disease recurrence. Furthermore, we review preliminary clinical data assessing the potential role of PARPi retreatment in patients who have previously progressed on PARPis.
Keywords: PARP inhibitors; drug resistance; ovarian neoplasms; platinum-based chemotherapy.
Conflict of interest statement
Author Mihaela Cristea was employed by the company Regeneron Pharmaceuticals. The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
Publication types
LinkOut - more resources
Full Text Sources

