The Effects of Lipid Extracts from Microalgae Chlorococcum amblystomatis and Nannochloropsis oceanica on the Proteome of 3D-Cultured Fibroblasts Exposed to UVA Radiation
- PMID: 40427427
- PMCID: PMC12108275
- DOI: 10.3390/antiox14050545
The Effects of Lipid Extracts from Microalgae Chlorococcum amblystomatis and Nannochloropsis oceanica on the Proteome of 3D-Cultured Fibroblasts Exposed to UVA Radiation
Abstract
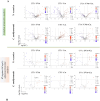
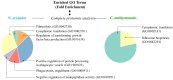
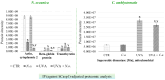
Nannochloropsis oceanica and Chlorococcum amblystomatis exhibit significant potential for protecting skin cells from oxidative stress-induced metabolic dysfunctions, owing to their high bioactive lipid content. This study aimed to evaluate their cytoprotective effects on the ultraviolet A (UVA)-perturbed proteome of 3D-cultured skin fibroblasts, using high-throughput proteomics. Chlorococcum amblystomatis lipid extract promoted a reduction in UVA-induced cytochrome c oxidase subunit 4 isoform 1 and cell death protein 6 levels, alongside the restoration of ferritin light chain expression diminished by UVA. It downregulated the expression of ubiquitin-conjugating enzyme E2 and lactoylglutathione lyase, which were upregulated by UVA. Furthermore, the elevated superoxide dismutase [Mn] mitochondrial levels in the caspase-1 interactome emphasized the lipid extract's role in mitigating oxidative stress-associated chronic inflammation by regulating caspase-1 activity. In addition to this notable redox balance-regulating and cytoprotective activity, conversely, the protein inflammation signaling mediated by UVA was regulated in terms of wound healing potential in the case of Nannochloropsis oceanica lipid extract. Following UVA radiation, it promoted the upregulation of complement component B, thrombospondin-1, MMP1, and fibulin-1. The results revealed that both lipid extracts effectively reversed the UVA-perturbed proteomic profile of fibroblasts, highlighting their therapeutic potential in protecting the skin from UV radiation.
Keywords: UVA radiation; freshwater microalgae Chlorococcum amblystomatis; human skin fibroblast; marine microalgae Nannochloropsis oceanica; proteomics; redox balance; wound healing.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures







Similar articles
-
Comparison of Microalgae Nannochloropsis oceanica and Chlorococcum amblystomatis Lipid Extracts Effects on UVA-Induced Changes in Human Skin Fibroblasts Proteome.Mar Drugs. 2024 Nov 10;22(11):509. doi: 10.3390/md22110509. Mar Drugs. 2024. PMID: 39590789 Free PMC article.
-
Comparison of the Regenerative Metabolic Efficiency of Lipid Extracts from Microalgae Nannochloropsis oceanica and Chlorococcum amblystomatis on Fibroblasts.Antioxidants (Basel). 2024 Feb 24;13(3):276. doi: 10.3390/antiox13030276. Antioxidants (Basel). 2024. PMID: 38539810 Free PMC article.
-
Impact of Nannochloropsis oceanica and Chlorococcum amblystomatis Extracts on UVA-Irradiated on 3D Cultured Melanoma Cells: A Proteomic Insight.Cells. 2024 Nov 21;13(23):1934. doi: 10.3390/cells13231934. Cells. 2024. PMID: 39682683 Free PMC article.
-
Polar Lipids of Marine Microalgae Nannochloropsis oceanica and Chlorococcum amblystomatis Mitigate the LPS-Induced Pro-Inflammatory Response in Macrophages.Mar Drugs. 2023 Dec 6;21(12):629. doi: 10.3390/md21120629. Mar Drugs. 2023. PMID: 38132950 Free PMC article.
-
Prevention of UVB Induced Metabolic Changes in Epidermal Cells by Lipid Extract from Microalgae Nannochloropsis oceanica.Int J Mol Sci. 2023 Jul 11;24(14):11302. doi: 10.3390/ijms241411302. Int J Mol Sci. 2023. PMID: 37511067 Free PMC article.
References
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

