Multiomics-Based Profiling of the Fecal Microbiome Reveals Potential Disease-Specific Signatures in Pediatric IBD (PIBD)
- PMID: 40427639
- PMCID: PMC12109367
- DOI: 10.3390/biom15050746
Multiomics-Based Profiling of the Fecal Microbiome Reveals Potential Disease-Specific Signatures in Pediatric IBD (PIBD)
Abstract
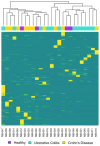
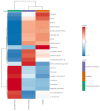
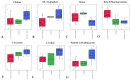
Inflammatory bowel disease (IBD), which includes Crohn's Disease (CD) and Ulcerative Colitis (UC), is a chronic gastrointestinal (GI) disorder affecting 1 in 100 people in the United States. Pediatric IBD (PIBD) is estimated to impact 15 per 100,000 children in North America. Factors such as the gut microbiome (GM), genetic predisposition to the disease, and certain environmental factors are thought to be involved in pathogenesis. However, the pathophysiology of IBD is incompletely understood, and diagnostic biomarkers and effective treatments, particularly for PIBD, are limited. Recent work suggests that these factors may interact to influence disease development, and multiomic approaches have emerged as promising tools to elucidate the pathophysiology. We employed metagenomics, metabolomics- and metatranscriptomics-based approaches to examine the microbiome, its genetic potential, and its activity to identify factors associated with PIBD. Metagenomics-based analyses revealed pathways such as octane oxidation and glycolysis that were differentially expressed in UC patients. Additionally, metatranscriptomics-based analyses suggested enrichment of glycan degradation and two component systems in UC samples as well as protein processing in the endoplasmic reticulum, ribosome, and protein export in CD and UC samples. In addition, metabolomics-based approaches revealed patterns of differentially abundant metabolites between healthy and PIBD individuals. Interestingly, overall microbiome community composition (as measured by alpha and beta diversity indices) did not appear to be associated with PIBD. However, we observed a small number of differentially abundant taxa in UC versus healthy controls, including members of the Classes Gammaproteobacteria and Clostridia as well as members of the Family Rikenellaceae. Accordingly, when identifying potential biomarkers for PIBD, our results suggest that multiomics-based approaches afford enhanced potential to detect putative biomarkers for PIBD compared to microbiome community composition sequence data alone.
Keywords: Crohn’s Disease; Ulcerative Colitis; biomarkers; inflammatory bowel disease; metabolomics; metagenomics; metatranscriptomics; microbiome; multiomics; pediatric inflammatory bowel disease.
Conflict of interest statement
The authors declare no conflicts of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures






References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

