Private Data Incrementalization: Data-Centric Model Development for Clinical Liver Segmentation
- PMID: 40428149
- PMCID: PMC12108710
- DOI: 10.3390/bioengineering12050530
Private Data Incrementalization: Data-Centric Model Development for Clinical Liver Segmentation
Abstract
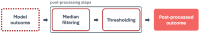
Machine Learning models, more specifically Artificial Neural Networks, are transforming medical imaging by enabling precise liver segmentation, a crucial task for diagnosing and treating liver diseases. However, these models often face challenges in adapting to diverse clinical data sources as differences in dataset volume, resolution, and origin impact generalization and performance. This study introduces a Private Data Incrementalization, a data-centric approach to enhance the adaptability of Artificial Neural Networks by progressively exposing them to varied clinical data. As the target of this study is not to propose a new image segmentation model, the existing medical imaging segmentation models-including U-Net, ResUNet++, Fully Convolutional Network, and a modified algorithm based on the Conditional Bernoulli Diffusion Model-are used. The study evaluates these four models using a curated private dataset of computed tomography scans from Coimbra University Hospital, supplemented by two public datasets, 3D-IRCADb01 and CHAOS. The Private Data Incrementalization method systematically increases the volume and diversity of training data, simulating real-world conditions where models must handle varied imaging contexts. Pre-processing and post-processing stages, incremental training, and performance evaluations reveal that structured exposure to diverse datasets improves segmentation performance, with ResUNet++ achieving the highest accuracy (0.9972) and Dice Similarity Coefficient (0.9449), and the best Average Symmetric Surface Distance (0.0053 mm), demonstrating the importance of dataset diversity and volume for segmentation models' robustness and generalization. Private Data Incrementalization thus offers a scalable strategy for building resilient segmentation models, ultimately benefiting clinical workflows, patient care, and healthcare resource management by addressing the variability inherent in clinical imaging data.
Keywords: BerDiff; FCN; ResUNet++; U-Net; artificial neural networks; automatic liver segmentation; computed tomography; private data incrementalization.
Conflict of interest statement
Author Ricardo Filipe was employed by the company Altice Labs, S.A. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
















References
-
- Batista S., Domingues I. Adoption of Artificial Neural Network-based Methods for Medical Image Segmentation; Proceedings of the Conferência Portuguesa de Reconhecimento de Padrões (RecPad); Covilhã, Portugal. 25 October 2024.
LinkOut - more resources
Full Text Sources
Research Materials

