Comparative Transcriptome Profiling of Ileal and Cecal Tissues Between Pekin Ducks and Shaoxing Ducks
- PMID: 40428310
- PMCID: PMC12110802
- DOI: 10.3390/genes16050488
Comparative Transcriptome Profiling of Ileal and Cecal Tissues Between Pekin Ducks and Shaoxing Ducks
Abstract
Background: Pekin ducks are well-known meat-type ducks, whereas Shaoxing ducks are bred for their egg-laying abilities. Growth and development of poultry species is well studied; however, very little is known regarding differences in intestinal gene expression between Pekin and Shaoxing ducks. Methods: To investigate intestinal differences between Pekin and Shaoxing ducks, we conducted transcriptome analysis on ileal and cecal tissues from five 42-day-old ducks per breed, raised under identical housing and feeding conditions to minimize environmental influences. Results: The results showed that a total of 379 differentially expressed genes (DEGs) with p < 0.05 and |log2FoldChange| > 1 were identified in the ileum when Pekin ducks were compared to Shaoxing ducks, among which 158 were upregulated and 221 were downregulated. Compared to Shaoxing ducks, a total of 367 DEGs with p < 0.05 and |log2FoldChange| > 1 were identified in the ceca of Pekin ducks, among which 204 were upregulated and 163 were downregulated. Among these DEGs, nine genes were reported to be associated with growth and metabolism, namely, P2RX6, KCNJ6, CASQ2, EHHADH, ACSBG1, ELOVL4, AIF1L, VILL, and FABP1. Functional enrichment analyses using the Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) databases indicated that the DEGs were significantly involved in pathways such as calcium signaling, unsaturated fatty acid biosynthesis, fatty acid degradation, and tryptophan metabolism. Conclusions: In conclusion, our study identified transcriptome differences in the intestines of meat-type and laying-type ducks, offering insights into the genetic basis of their growth and metabolic differences. Future studies should validate key genes and explore environmental influences on gene expression.
Keywords: cecum; duck; ileum; transcriptome.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures



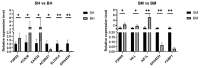
Similar articles
-
Transcriptome Analysis Reveals the Genes Involved in Growth and Metabolism in Muscovy Ducks.Biomed Res Int. 2021 Apr 17;2021:6648435. doi: 10.1155/2021/6648435. eCollection 2021. Biomed Res Int. 2021. PMID: 33959661 Free PMC article.
-
Changes in morphology and miRNAs expression in small intestines of Shaoxing ducks in response to high temperature.Mol Biol Rep. 2019 Aug;46(4):3843-3856. doi: 10.1007/s11033-019-04827-2. Epub 2019 May 2. Mol Biol Rep. 2019. PMID: 31049835
-
Comparative Transcriptome Profiling of Ovary Tissue between Black Muscovy Duck and White Muscovy Duck with High- and Low-Egg Production.Genes (Basel). 2020 Dec 31;12(1):57. doi: 10.3390/genes12010057. Genes (Basel). 2020. PMID: 33396489 Free PMC article.
-
Characterization and comparative transcriptomic analysis of skeletal muscle in female Pekin duck and Hanzhong Ma duck during different growth stages using RNA-seq.Poult Sci. 2023 Dec;102(12):103122. doi: 10.1016/j.psj.2023.103122. Epub 2023 Sep 18. Poult Sci. 2023. PMID: 37832186 Free PMC article.
-
Dynamic accumulation of fatty acids in duck (Anas platyrhynchos) breast muscle and its correlations with gene expression.BMC Genomics. 2020 Jan 17;21(1):58. doi: 10.1186/s12864-020-6482-7. BMC Genomics. 2020. PMID: 31952469 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

