Integrated Genomic and Transcriptomic Analysis Reveals a Transcription Factor Gene Set Facilitating Gonadal Differentiation in the Pacific Oyster Crassostrea gigas
- PMID: 40428334
- PMCID: PMC12111275
- DOI: 10.3390/genes16050513
Integrated Genomic and Transcriptomic Analysis Reveals a Transcription Factor Gene Set Facilitating Gonadal Differentiation in the Pacific Oyster Crassostrea gigas
Abstract
Background/objectives: The Pacific oyster Crassostrea gigas has emerged as a promising model system for sex determination studies due to its complex reproduction strategy and sex reversal. Transcription factors (TFs) play crucial roles in sex determination and gonadal differentiation. Despite previous research revealing functions of several conserved sex-determining pathway genes, such as Dmrt1, Foxl2, and SoxH, little is known about the other essential TF regulators driving C. gigas gonadal differentiation and development.
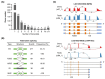
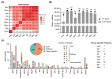
Methods: In this study, a systematic identification of TFs revealed 1167 TF genes in the C. gigas genome. Comparative transcriptome analysis of C. gigas female and male gonads demonstrated 123 differentially expressed TF genes.
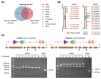
Results: The majority of these sex-related TF genes were up-regulated in female or male gonads from the inactive stage to the mature stage. Moreover, this TF gene set was deeply conserved and showed similar regulation in the Kumamoto oyster Crassostrea sikamea gonads, suggesting their important regulatory roles in gonadal differentiation and development in Crassostrea oysters. Furthermore, two BTB TF gene clusters were identified in the C. gigas genome, both of which were specifically expressed in the male gonad. Gene numbers of each BTB gene cluster showed significant variations among six Crassostrea species.
Conclusions: To the best of our knowledge, this study provides the first report of the whole TF family in C. gigas. The sex-related TF gene set will be a valuable resource for further research aimed at uncovering TF gene regulatory networks in oyster sex determination and gonadal differentiation.
Keywords: Crassostrea gigas; Pacific oyster; gonadal differentiation; sex determination; transcription factor.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





Similar articles
-
Gonad Transcriptome Analysis of the Pacific Oyster Crassostrea gigas Identifies Potential Genes Regulating the Sex Determination and Differentiation Process.Mar Biotechnol (NY). 2018 Apr;20(2):206-219. doi: 10.1007/s10126-018-9798-4. Epub 2018 Mar 8. Mar Biotechnol (NY). 2018. PMID: 29516375
-
Variance in expression and localization of sex-related genes CgDsx, CgBHMG1 and CgFoxl2 during diploid and triploid Pacific oyster Crassostrea gigas gonad differentiation.Gene. 2021 Jul 20;790:145692. doi: 10.1016/j.gene.2021.145692. Epub 2021 May 4. Gene. 2021. PMID: 33961972
-
Genomic analysis of the Pacific oyster (Crassostrea gigas) reveals possible conservation of vertebrate sex determination in a mollusc.G3 (Bethesda). 2014 Sep 11;4(11):2207-17. doi: 10.1534/g3.114.013904. G3 (Bethesda). 2014. PMID: 25213692 Free PMC article.
-
Comprehensive analysis of Sox genes in the Pacific oyster (Crassostrea gigas): Insights into expression and potential functions.Comp Biochem Physiol Part D Genomics Proteomics. 2025 Sep;55:101463. doi: 10.1016/j.cbd.2025.101463. Epub 2025 Mar 3. Comp Biochem Physiol Part D Genomics Proteomics. 2025. PMID: 40147269
-
Sex determination and gonadal sex differentiation in the chicken model.Int J Dev Biol. 2018;62(1-2-3):153-166. doi: 10.1387/ijdb.170319cs. Int J Dev Biol. 2018. PMID: 29616724 Review.
Cited by
-
Unveiling the Bioactive Potential of the Invasive Jellyfish Phyllorhiza punctata Through Integrative Transcriptomic and Proteomic Analyses.Biomolecules. 2025 Aug 4;15(8):1121. doi: 10.3390/biom15081121. Biomolecules. 2025. PMID: 40867566 Free PMC article.
References
-
- Santerre C., Sourdaine P., Marc N., Mingant C., Robert R., Martinez A.-S. Oyster sex determination is influenced by temperature—First clues in spat during first gonadic differentiation and gametogenesis. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 2013;165:61–69. doi: 10.1016/j.cbpa.2013.02.007. - DOI - PubMed
-
- Dang X., Zhang Y., Dupont S., Gaitán-Espitia J.D., He Y.-Q., Wang H.-H., Ellis R.P., Guo X., Parker L., Zhang R.-C., et al. Low pH means more female offspring: A multigenerational plasticity in the sex ratio of marine bivalves. Environ. Sci. Technol. 2024;59:1308–1321. doi: 10.1021/acs.est.4c07808. - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

