Genome-Wide Association Studies and Candidate Genes for Egg Production Traits in Layers from an F2 Crossbred Population Produced Using Two Divergently Selected Chicken Breeds, Russian White and Cornish White
- PMID: 40428405
- PMCID: PMC12111742
- DOI: 10.3390/genes16050583
Genome-Wide Association Studies and Candidate Genes for Egg Production Traits in Layers from an F2 Crossbred Population Produced Using Two Divergently Selected Chicken Breeds, Russian White and Cornish White
Abstract
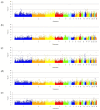
Background/Objectives: Finding single nucleotide polymorphisms (SNPs) and candidate genes that influence the expression of key traits is essential for genomic selection and helps improve the efficiency of poultry production. Here, we aimed to conduct a genome-wide association study (GWAS) for egg production traits in an F2 resource population of chickens (Gallus gallus). Methods: The examined F2 population was produced by crossing two divergently selected breeds with contrasting phenotypes for egg performance traits, namely Russian White (of higher egg production) and Cornish White (of lower egg production). Sampled birds (n = 142) were genotyped using the Illumina Chicken 60K SNP iSelect BeadChip. Results: In the course of the GWAS analysis, we were able to clarify significant associations with phenotypic traits of interest and economic value by using 47,432 SNPs after the genotype dataset was filtered. At the threshold p < 1.06 × 10-6, we found 23 prioritized candidate genes (PCGs) associated with egg weight at the age of 42-52 weeks (FGF14, GCK), duration of egg laying (CNTN4), egg laying cycle (SAMD12) and egg laying interval (PHF5A, AKR1B1, CALD1, ATP7B, PIK3R4, PTK2, PRKCE, FAT1, PCM1, CC2D2A, BMS1, SEMA6D, CDH13, SLIT3, ATP10B, ISCU, LRRC75A, LETM2, ANKRD24). Moreover, two SNPs were co-localized within the FGF14 gene. Conclusions: Based on our GWAS findings, the revealed SNPs and candidate genes can be used as genetic markers for egg weight and other performance characteristics in chickens to attain genetic enhancement in production and for further genomic selection.
Keywords: candidate genes; chicken (Gallus gallus); egg performance; genome-wide association study (GWAS); single nucleotide polymorphisms (SNPs).
Conflict of interest statement
The authors declare no conflicts of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures



References
-
- Meuwissen T., Hayes B., MacLeod I., Goddard M. Identification of genomic variants causing variation in quantitative traits: A review. Agriculture. 2022;12:1713. doi: 10.3390/agriculture12101713. - DOI
-
- Plemyashov K.V., Smaragdov M.G., Romanov M.N. Molecular Genetic Technologies for Analysis of Gene Expression Related to Animal Productivity and Disease Resistance, Proceedings of the Materials of the 3rd International Scientific and Practical Conference, Moscow, Russia, 30 September 2021. Sel’skokhozyaistvennye Tekhnologii; Moscow, Russia: 2021. Molekulyarno-geneticheskiy polimorfizm v populyatsiyakh zhivotnykh i yego primeneniye v intensivnoy selektsii molochnogo skota: Obzor [Molecular Genetic Polymorphism in Animal Populations and Its Application in Intensive Breeding of Dairy Cattle—A Review] pp. 368–378. (In Russian with English summary)
-
- Plemyashov K.V., Smaragdov M.G., Romanov M.N. Genomnaya otsenka plemennykh bykov [Genomic Assessment of Breeding Bulls] In: Pozyabin S.V., Kochish I.I., Romanov M.N., editors. Molecular Genetic Technologies for Analysis of Gene Expression Related to Animal Productivity and Disease Resistance, Proceedings of the Materials of the 3rd International Scientific and Practical Conference, Moscow, Russia, 30 September 2021. Sel’skokhozyaistvennye Tekhnologii; Moscow, Russia: 2021. [(accessed on 21 April 2025)]. pp. 363–367. Available online: https://elib.spbstu.ru/dl/2/z21-43.pdf/info.
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

