LRR Receptor-like Protein in Rapeseed Confers Resistance to Sclerotinia sclerotiorum Infection via a Conserved Ss NEP2 Peptide
- PMID: 40429714
- PMCID: PMC12110989
- DOI: 10.3390/ijms26104569
LRR Receptor-like Protein in Rapeseed Confers Resistance to Sclerotinia sclerotiorum Infection via a Conserved Ss NEP2 Peptide
Abstract
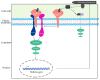
Brassica napus is one of the most extensively cultivated oilseed crops in China, but its yield is significantly impacted by stem rot caused by Sclerotinia sclerotiorum. Receptor-like proteins (RLPs) and receptor-like kinases (RLKs) play essential roles in plant-pathogen interactions; however, their regulatory mechanisms remain largely unknown in B. napus. In this study, we investigated the function of the leucine-rich repeat receptor-like protein BnaRLP-G13-1 in Brassica napus immunity. Previous observations indicated that B. napus plants expressing BnaRLP-G13-1 exhibited enhanced resistance to Sclerotinia sclerotiorum. We hypothesized that BnaRLP-G13-1 mediates pathogen recognition and immune signaling. To test this, we employed mitogen-activated protein kinase (MAPK) activity assays, transgenic overexpression analyses, and pathogen infection assays. Our results demonstrated that BnaRLP-G13-1 recognizes the conserved necrosis- and ethylene-inducing peptide Ssnlp24SsNEP2 derived from S. sclerotiorum, triggering MAPK cascades and subsequent immune responses. Furthermore, protein interaction studies revealed that BnaRLP-G13-1 physically interacts with the receptor-like kinase BnaSOBIR1, which is essential for full antifungal defense activation. These results elucidate the molecular basis of BnaRLP-G13-1-mediated immunity, providing insights into improving disease resistance in oilseed crops.
Keywords: BnaSOBIR1; Brassica napus; LRR-RLP; Sclerotinia sclerotiorum; SsNEP2; plant immunity.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures







Similar articles
-
A GDSL motif-containing lipase modulates Sclerotinia sclerotiorum resistance in Brassica napus.Plant Physiol. 2024 Dec 2;196(4):2973-2988. doi: 10.1093/plphys/kiae500. Plant Physiol. 2024. PMID: 39321167 Free PMC article.
-
Comparative transcriptomic analysis uncovers the complex genetic network for resistance to Sclerotinia sclerotiorum in Brassica napus.Sci Rep. 2016 Jan 8;6:19007. doi: 10.1038/srep19007. Sci Rep. 2016. PMID: 26743436 Free PMC article.
-
Overexpression of OsPGIP2 confers Sclerotinia sclerotiorum resistance in Brassica napus through increased activation of defense mechanisms.J Exp Bot. 2018 May 25;69(12):3141-3155. doi: 10.1093/jxb/ery138. J Exp Bot. 2018. PMID: 29648614 Free PMC article.
-
Ceramide-1-phosphate enhances defense responses against Sclerotinia sclerotiorum in Brassica napus.Plant Physiol. 2025 Feb 7;197(2):kiae649. doi: 10.1093/plphys/kiae649. Plant Physiol. 2025. PMID: 39928582
-
Sclerotinia Stem Rot Resistance in Rapeseed: Recent Progress and Future Prospects.J Agric Food Chem. 2021 Mar 17;69(10):2965-2978. doi: 10.1021/acs.jafc.0c07351. Epub 2021 Mar 5. J Agric Food Chem. 2021. PMID: 33667087 Review.
References
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources

