Genome-Wide Characterization of WRKY Gene Family in Camellia chekiangoleosa Identifies Potential Regulatory Components in Pigment Biosynthesis Pathways
- PMID: 40429766
- PMCID: PMC12111399
- DOI: 10.3390/ijms26104622
Genome-Wide Characterization of WRKY Gene Family in Camellia chekiangoleosa Identifies Potential Regulatory Components in Pigment Biosynthesis Pathways
Abstract
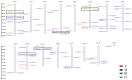
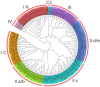
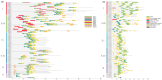
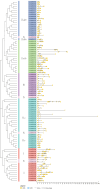
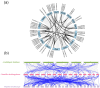
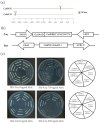
The WRKY gene family is essential for controlling a variety of plant physiological functions, yet the involvement of specific WRKY members in pigment biosynthesis and accumulation in Camellia chekiangoleosa remains unexplored, particularly in anthocyanins and carotenoids, which play crucial roles in the pigmentation of C. chekiangoleosa. This study systematically identified 87 WRKY genes across 15 chromosomes in C. chekiangoleosa through bioinformatic approaches. Further structural and phylogenetic analyses of these TFs enabled their classification into six different subgroups. WRKY family expansion was shown to be mostly driven by tandem duplication. W-box elements, which can be binding sites for WRKY transcription factors, were present in a number of biosynthetic genes in the pigment production pathway. Yeast one-hybrid assay confirmed that five WRKY transcription factors (CchWRKY15/24/33/47/76) directly bind to the promoter regions of two key biosynthetic genes, CchPSY1 and Cch4CL1. Intriguingly, among the five WRKYs tested, the expression levels of CchWRKY15, CchWRKY33, and CchWRKY47 showed the strongest positive associations with flavonoid accumulation (p < 0.05, Pearson correlation analysis).These findings provide novel insights into the evolutionary patterns, transcriptional regulation, and functional characteristics of CchWRKYs, while elucidating their possible regulatory mechanisms in the fruit coloration of C. chekiangoleosa.
Keywords: Camellia chekiangoleosa; WRKY gene family; fruit coloration; pigment synthesis; transcriptional regulation.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures



Similar articles
-
Gene Structural Specificity and Expression of MADS-Box Gene Family in Camellia chekiangoleosa.Int J Mol Sci. 2023 Feb 8;24(4):3434. doi: 10.3390/ijms24043434. Int J Mol Sci. 2023. PMID: 36834845 Free PMC article.
-
Cytological, Physiological, and Transcriptome Analysis of Leaf-Yellowing Mutant in Camellia chekiangoleosa.Int J Mol Sci. 2024 Dec 27;26(1):132. doi: 10.3390/ijms26010132. Int J Mol Sci. 2024. PMID: 39795989 Free PMC article.
-
Genome-wide identification of WRKY family genes and their response to abiotic stresses in tea plant (Camellia sinensis).Genes Genomics. 2019 Jan;41(1):17-33. doi: 10.1007/s13258-018-0734-9. Epub 2018 Sep 20. Genes Genomics. 2019. PMID: 30238224
-
Genome-Wide Identification and Expression Analysis of WRKY Transcription Factors under Multiple Stresses in Brassica napus.PLoS One. 2016 Jun 20;11(6):e0157558. doi: 10.1371/journal.pone.0157558. eCollection 2016. PLoS One. 2016. PMID: 27322342 Free PMC article.
-
Unraveling the involvement of WRKY TFs in regulating plant disease defense signaling.Planta. 2023 Nov 28;259(1):7. doi: 10.1007/s00425-023-04269-y. Planta. 2023. PMID: 38012461 Review.
References
MeSH terms
Substances
Grants and funding
- 2021YFD1000402/National Key Research and Development Program of China
- 32201592/National Natural Science Foundation of China
- 20232BAB215045 and 20242BAB20305/Natural Science Foundation of Jiangxi Province, China
- 20224BBF61027/Key Research and Development Program of Jiangxi Province
- 20212BCD46002/Science and Technology Innovation Bases Program of Jiangxi province, China
- YCYJZX [2023] 131and YCYJZX [2023] 111/Oil-tea special research project of Jiangxi Provincial Department of Forestry
- 202228/WT_/Wellcome Trust/United Kingdom
- 20244BCE52294/Jiangxi Province Early-Career Young Scientific and Technological Talent Development Found
- 2025521002/Special Project for Basic Research and Technology Transfer and Transformation of the Jiangxi Academy of Forestry
LinkOut - more resources
Full Text Sources

