Genetic, Transcriptomic, and Epigenomic Insights into Sjögren's Disease: An Integrative Network Investigation and Immune Diseases Comparison
- PMID: 40429780
- PMCID: PMC12111751
- DOI: 10.3390/ijms26104637
Genetic, Transcriptomic, and Epigenomic Insights into Sjögren's Disease: An Integrative Network Investigation and Immune Diseases Comparison
Abstract
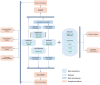
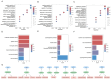
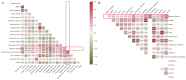
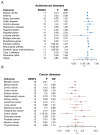
Sjögren's disease (SjD) is a systemic autoimmune disorder primarily causing dry eyes and mouth. It frequently overlaps with other autoimmune diseases (AIDs), including rheumatoid arthritis (RA) and systemic lupus erythematosus (SLE). However, the genetic basis of SjD remains underexplored, limiting our understanding of its connections to other immune-mediated conditions. In this study, we aimed to identify gene networks associated with SjD through the integration of genetic, transcriptomic, and epigenomic data. We further compared the genetic factors of SjD with other immune-mediated diseases. We analyzed genome-wide association studies (GWAS) summary statistics, DNA methylation, and transcriptomic data using our in-house network-based methods, dmGWAS and EW_dmGWAS, to identify key gene modules associated with SjD. In dmGWAS analysis, discovery and evaluation datasets were used to identify consensus results. We conducted gene-set, cell-type, and disease-enrichment analyses on significant gene modules and explored potential drug targets. Genetic correlations and Mendelian randomization were applied to assess SjD's link with 17 other AIDs and 16 cancer types. dmGWAS identified 207 and 211 gene modules in the discovery and evaluation phases, respectively, while EW_dmGWAS detected 886 modules. Key modules highlighted 55 genes (discovery), 52 genes (evaluation), and 59 genes (EW_dmGWAS), with at least 50 genes from each analysis linked to AIDs and cancer. Enrichment analyses confirmed their relevance to immune and oncogenic pathways. We pinpointed four candidate drug targets associated with AIDs. We developed a novel integrative omics approach to identify potential genetic markers of SjD and compared them with AIDs and cancers. Our approach can be similarly applied to other disease studies.
Keywords: Mendelian randomization; autoimmune disease; drug target; genetic correlation; genome-wide association studies; pleiotropy.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

Similar articles
-
GeneHarmony: A Knowledge-Based Tool for Biomarker Discovery in Disease: Sjögren's Disease vs. Rheumatoid Arthritis and Systemic Lupus Erythematosus.Int J Mol Sci. 2025 Jul 2;26(13):6379. doi: 10.3390/ijms26136379. Int J Mol Sci. 2025. PMID: 40650157 Free PMC article.
-
Multi-omics analysis for identifying cell-type-specific and bulk-level druggable targets in Alzheimer's disease.J Transl Med. 2025 Jul 13;23(1):788. doi: 10.1186/s12967-025-06739-1. J Transl Med. 2025. PMID: 40653482 Free PMC article.
-
A high polygenic risk score is associated with SSA/SSB antibody positivity and early onset in primary Sjögren's disease.Rheumatology (Oxford). 2025 Jul 1;64(7):4341-4346. doi: 10.1093/rheumatology/keae693. Rheumatology (Oxford). 2025. PMID: 39693120 Free PMC article.
-
Interventions for the management of dry mouth: non-pharmacological interventions.Cochrane Database Syst Rev. 2013 Sep 5;2013(9):CD009603. doi: 10.1002/14651858.CD009603.pub3. Cochrane Database Syst Rev. 2013. PMID: 24006231 Free PMC article.
-
Systemic pharmacological treatments for chronic plaque psoriasis: a network meta-analysis.Cochrane Database Syst Rev. 2021 Apr 19;4(4):CD011535. doi: 10.1002/14651858.CD011535.pub4. Cochrane Database Syst Rev. 2021. Update in: Cochrane Database Syst Rev. 2022 May 23;5:CD011535. doi: 10.1002/14651858.CD011535.pub5. PMID: 33871055 Free PMC article. Updated.
References
-
- Kuo C.F., Grainge M.J., Valdes A.M., See L.C., Luo S.F., Yu K.H., Zhang W., Doherty M. Familial Risk of Sjogren’s Syndrome and Co-aggregation of Autoimmune Diseases in Affected Families: A Nationwide Population Study. Arthritis Rheumatol. 2015;67:1904–1912. doi: 10.1002/art.39127. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

