Computational Search for Inhibitors of SOD1 Mutant Infectivity as Potential Therapeutics for ALS Disease
- PMID: 40429802
- PMCID: PMC12111112
- DOI: 10.3390/ijms26104660
Computational Search for Inhibitors of SOD1 Mutant Infectivity as Potential Therapeutics for ALS Disease
Abstract
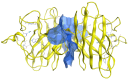
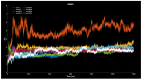
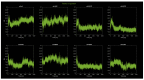
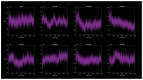
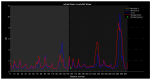
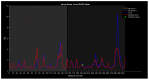
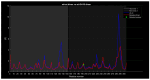
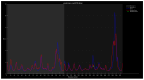
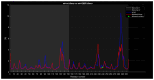
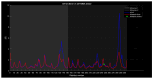
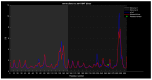
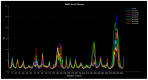
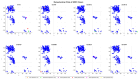
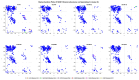
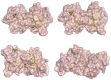
Familial amyotrophic lateral sclerosis (ALS) is a progressive neurodegenerative disease characterized by the selective degeneration of motor neurons. Among the main genetic causes of ALS, over 200 mutations have been identified in the Cu/Zn superoxide dismutase (SOD1) protein, a dimeric metalloenzyme essential for converting superoxides from cellular respiration into less toxic products. Point mutations in SOD1 monomers can induce protein misfolding, which spreads to wild-type monomers through a prion-like mechanism, leading to dysfunctions that contribute to the development of the disease. Understanding the structural and functional differences between the wild-type protein and its mutated variants, as well as developing drugs capable of inhibiting the propagation of misfolding, is crucial for identifying new therapeutic strategies. In this work, seven SOD1 mutations (A4V, G41D, G41S, D76V, G85R, G93A, and I104F) were selected, and three-dimensional models of SOD1 dimers composed of one wild-type monomer and one mutated monomer were generated, along with a control dimer consisting solely of wild-type monomers. Molecular dynamics simulations were conducted to investigate conformational differences between the dimers. Additionally, molecular docking was performed using a library of ligands to identify compounds with high affinity for the mutated dimers. The study reveals some differences in the mutated dimers following molecular dynamics simulations and in the docking of the selected ligands with the various dimers.
Keywords: ALS mutations; SOD1 inhibitors; docking; molecular dynamics.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures
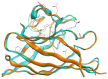

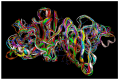



References
-
- Brotman R.G., Moreno-Escobar M.C., Joseph J., Munakomi S., Pawar G. Amyotrophic Lateral Sclerosis. StatPearls; Treasure Island, FL, USA: 2025. - PubMed
-
- Bernard E., Pegat A., Svahn J., Bouhour F., Leblanc P., Millecamps S., Thobois S., Guissart C., Lumbroso S., Mouzat K. Clinical and molecular landscape of ALS patients with SOD1 mutations: Novel pathogenic variants and novel phenotypes. A single ALS center study. Int. J. Mol. Sci. 2020;21:6807. doi: 10.3390/ijms21186807. - DOI - PMC - PubMed
-
- ALSoD. [(accessed on 1 April 2024)]. Available online: https://alsod.ac.uk/output/gene.php/SOD1.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

