Transcriptional Regulatory Systems in Pseudomonas: A Comparative Analysis of Helix-Turn-Helix Domains and Two-Component Signal Transduction Networks
- PMID: 40429820
- PMCID: PMC12112638
- DOI: 10.3390/ijms26104677
Transcriptional Regulatory Systems in Pseudomonas: A Comparative Analysis of Helix-Turn-Helix Domains and Two-Component Signal Transduction Networks
Abstract
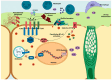
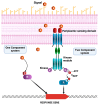
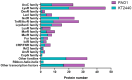
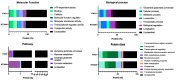
Bacterial communities in diverse environmental niches respond to various external stimuli for survival. A primary means of communication between bacterial cells involves one-component (OC) and two-component signal transduction systems (TCSs). These systems are key for sensing environmental changes and regulating bacterial physiology. TCSs, which are the more complex of the two, consist of a sensor histidine kinase for receiving an external input and a response regulator to convey changes in bacterial cell physiology. For numerous reasons, TCSs have emerged as significant targets for antibacterial drug design due to their role in regulating expression level, bacterial viability, growth, and virulence. Diverse studies have shown the molecular mechanisms by which TCSs regulate virulence and antibiotic resistance in pathogenic bacteria. In this study, we performed a thorough analysis of the data from multiple public databases to assemble a comprehensive catalog of the principal detection systems present in both the non-pathogenic Pseudomonas putida KT2440 and the pathogenic Pseudomonas aeruginosa PAO1 strains. Additionally, we conducted a sequence analysis of regulatory elements associated with transcriptional proteins. These were classified into regulatory families based on Helix-turn-Helix (HTH) protein domain information, a common structural motif for DNA-binding proteins. Moreover, we highlight the function of bacterial TCSs and their involvement in functions essential for bacterial survival and virulence. This comparison aims to identify novel targets that can be exploited for the development of advanced biotherapeutic strategies, potentially leading to new treatments for bacterial infections.
Keywords: Pseudomonas aeruginosa PAO1; Pseudomonas putida KT2440; gene expression; metabolism; one-component system (OCS); pathogenicity; two-component system (TCS).
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures


Similar articles
-
Two-Component Signal Transduction Systems of Pathogenic Bacteria As Targets for Antimicrobial Therapy: An Overview.Front Microbiol. 2017 Oct 10;8:1878. doi: 10.3389/fmicb.2017.01878. eCollection 2017. Front Microbiol. 2017. PMID: 29067003 Free PMC article. Review.
-
Role of Two-Component System Networks in Pseudomonas aeruginosa Pathogenesis.Adv Exp Med Biol. 2022;1386:371-395. doi: 10.1007/978-3-031-08491-1_14. Adv Exp Med Biol. 2022. PMID: 36258080
-
Functional and Pangenomic Exploration of Roc Two-Component Regulatory Systems Identifies Novel Players Across Pseudomonas Species.Mol Microbiol. 2025 May;123(5):439-453. doi: 10.1111/mmi.15357. Epub 2025 Mar 14. Mol Microbiol. 2025. PMID: 40087830 Free PMC article.
-
Roles of Two-Component Signal Transduction Systems in Shigella Virulence.Biomolecules. 2022 Sep 18;12(9):1321. doi: 10.3390/biom12091321. Biomolecules. 2022. PMID: 36139160 Free PMC article. Review.
-
Insights into the atypical autokinase activity of the Pseudomonas aeruginosa GacS histidine kinase and its interaction with RetS.Structure. 2022 Sep 1;30(9):1285-1297.e5. doi: 10.1016/j.str.2022.06.002. Epub 2022 Jun 28. Structure. 2022. PMID: 35767996
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

