Helix Folding in One Dimension: Effects of Proline Co-Solvent on Free Energy Landscape of Hydrogen Bond Dynamics in Alanine Peptides
- PMID: 40430235
- PMCID: PMC12113030
- DOI: 10.3390/life15050809
Helix Folding in One Dimension: Effects of Proline Co-Solvent on Free Energy Landscape of Hydrogen Bond Dynamics in Alanine Peptides
Abstract
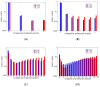
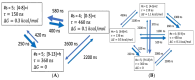
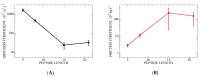
The effects of proline co-solvent on helix folding are explored through the single discrete coordinate of the number of helical hydrogen bonds. The analysis is based on multi-microsecond length molecular dynamics simulations of alanine-based helix-forming peptides, (ALA)n, of length n = 4, 8, 15 and 21 residues, in an aqueous solution with 2 M concentration of proline. The effects of addition of proline on the free energy landscape for helix folding were analyzed using the graph-based Dijkstra algorithm, Optimal Dimensionality Reduction kinetic coarse graining, committor functions, as well as through the diffusion of the helix boundary. Viewed at a sufficiently long time-scale, helix folding in the coarse-grained hydrogen bond space follows a consecutive mechanism, with well-defined initiation and propagation phases, and an interesting set of intermediates. Proline addition slows down the folding relaxation of all four peptides, increases helix content and induces subtle mechanistic changes compared to pure water solvation. A general trend is for transition state shift towards earlier stages of folding in proline relative to water. For ALA5 and ALA8 direct folding is dominant. In ALA8 and ALA15 multiple pathways appear possible. For ALA21 a simple mechanism emerges, with a single path from helix to coil through a set of intermediates. Overall, this work provides new insights into effects of proline co-solvent on helix folding, complementary to more standard approaches based on three-dimensional molecular structures.
Keywords: alanine peptides; helix folding; hydrogen bond dynamics; proline co-solvent.
Conflict of interest statement
The authors declare no conflict of interest.
Figures







Similar articles
-
Microscopic effects of proline co-solvent on alanine homopeptide structure, solvation and helix folding dynamics.J Biomol Struct Dyn. 2025 May 12:1-11. doi: 10.1080/07391102.2025.2500681. Online ahead of print. J Biomol Struct Dyn. 2025. PMID: 40351163
-
Length Dependent Folding Kinetics of Alanine-Based Helical Peptides from Optimal Dimensionality Reduction.Life (Basel). 2021 Apr 24;11(5):385. doi: 10.3390/life11050385. Life (Basel). 2021. PMID: 33923197 Free PMC article.
-
Effects of Proline on Internal Friction in Simulated Folding Dynamics of Several Alanine-Based α-Helical Peptides.J Phys Chem B. 2024 Apr 25;128(16):3856-3869. doi: 10.1021/acs.jpcb.4c00623. Epub 2024 Apr 12. J Phys Chem B. 2024. PMID: 38606880 Free PMC article.
-
Helix-Coil Transition Courses Through Multiple Pathways and Intermediates: Fast Kinetic Measurements and Dimensionality Reduction.J Phys Chem B. 2018 Dec 6;122(48):10806-10816. doi: 10.1021/acs.jpcb.8b07924. Epub 2018 Nov 27. J Phys Chem B. 2018. PMID: 30395709
-
Scrutiny of chain-length and N-terminal effects in α-helix folding: a molecular dynamics study on polyalanine peptides.J Biomol Struct Dyn. 2017 Jul;35(9):1923-1935. doi: 10.1080/07391102.2016.1199972. Epub 2016 Jul 6. J Biomol Struct Dyn. 2017. PMID: 27310440
References
-
- Kuriyan J., Konforti B., Wemmer D. The Molecules of Life. Garland Science; New York, NY, USA: 2013.
LinkOut - more resources
Full Text Sources

