Identification of a Papain-like Cysteine Protease Functioning as an Avirulence Factor in Striga-Cowpea Interactions
- PMID: 40430992
- PMCID: PMC12114844
- DOI: 10.3390/plants14101427
Identification of a Papain-like Cysteine Protease Functioning as an Avirulence Factor in Striga-Cowpea Interactions
Abstract
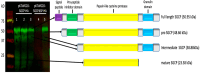
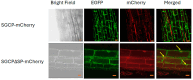
While most cowpea cultivars are susceptible to parasitism by the root parasitic weed Striga gesnerioides (Willd.) Vatke, cultivar B301 is resistant to all Striga races except for SG4z. Resistance to Striga parasitism is manifested by the elicitation of a hypersensitive response (HR) at the site of parasite attachment on the host root followed by rapid death of the attached parasite. We isolated a papain-like cysteine protease (PLCP) designated SGCP1 that is highly expressed in the haustoria of S. gesnerioides race SG3 at the time of parasite attachment to the host root. SGCP1 contains an apoplast-targeting signal peptide, a Cathepsin pro-peptide inhibitory domain, a papain family cysteine protease domain, and a granulin domain. Full-length SGCP1 and a variant lacking the signal peptide (SGCP∆SP) were expressed in the roots of composite B301 plants. Expression of SGCP1 and SGCP∆SP resulted in activation of host innate immune responses exemplified by increased frequency of HR and decreased levels of parasite cotyledon expansion (CE), indicative of successful host parasitism, in transgenic compared to wild-type B301 roots parasitized by SG4z. These data indicate that SGCP1 functions as an avirulence factor capable of activating host innate immunity and furthers our understanding of how compatible and incompatible host-parasite interactions are controlled.
Keywords: Striga gesnerioides; avirulence; cowpea; cysteine protease; innate immunity.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures





References
-
- Parker C. Parasitic Orobanchaceae: Parasitic Mechanisms and Control Strategies. Springer; Berlin/Heidelberg, Germany: 2013. The parasitic weeds of the Orobanchaceae; pp. 313–344.
-
- Barberi P. Ecological weed management in sub-Saharan Africa: Prospects and implications on other agroecosystem services. Adv. Agron. 2019;156:219–264.
-
- Timko M.P., Singh B.B. Genomics of Tropical Crop Plants. Springer; New York, NY, USA: 2008. Chapter 10. Cowpea, a multifunctional legume; pp. 227–258. Plant Genetics and Genomics: Crops and Models.
Grants and funding
LinkOut - more resources
Full Text Sources

