Identification and Expression Analysis of NAC Transcription Factors Related to Rust Resistance in Foxtail Millet
- PMID: 40431072
- PMCID: PMC12115137
- DOI: 10.3390/plants14101507
Identification and Expression Analysis of NAC Transcription Factors Related to Rust Resistance in Foxtail Millet
Abstract
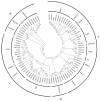
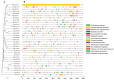
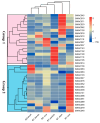
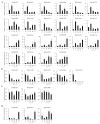
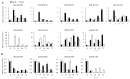
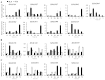
Foxtail millet (Setaria italica), a vital cereal crop in China, serves as both a staple food and forage source but is threatened by rust disease caused by Uromyces setariae-italicae (Usi), leading to severe yield and quality losses. The NAM, ATAF1/2, and CUC2 (NAC) transcription factor family represents one of the largest plant-specific regulatory gene families, playing pivotal roles in development and stress responses. However, the functional relevance of NAC genes in foxtail millet's defense against this pathogen remains unexplored. Here, we systematically analyzed 33 SiNAC genes from the foxtail millet genome. Phylogenetic analysis classified these genes into 11 subgroups, while chromosomal mapping localized them to nine chromosomes unevenly. Promoter analysis identified stress- and plant hormone-related cis-elements, suggesting functional diversity. Expression profiling analysis showed that most SiNAC genes exhibit tissue-specific expression patterns. Quantitative real-time PCR (qRT-PCR) results indicated that 30 genes responded to Usi infection, with 17 showing a strong association with rust resistance. Three resistance-associated genes demonstrated transactivation activity and nuclear localization, indicating their regulatory function in defense responses. This study provides both mechanistic insights into SiNAC-mediated rust resistance and potential targets for molecular breeding in foxtail millet.
Keywords: NAC; Setaria italica; Uromyces setariae-italicae; qRT-PCR; rust disease resistance; transcription factor.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures



References
-
- Liu J.S., Shao L.X., Cui G.X., Zhang H.Q. A preliminary study on the epidemic characteristics of millet rust. J. Agric. Univ. Hebei. 1997;20:30–34. (In Chinese)
-
- Shan W., Kuang J.F., Chen L., Xie H., Peng H.H., Xiao Y.Y., Li X.P., Chen W.X., He Q.G., Chen J.Y., et al. Molecular characterization of banana NAC transcription factors and their interactions with ethylene signalling component EIL during fruit ripening. J. Exp. Bot. 2012;63:5171–5187. doi: 10.1093/jxb/ers178. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

