Investigating Skin Microbial Community in Malignant Melanoma Lesions
- PMID: 40431165
- PMCID: PMC12113796
- DOI: 10.3390/microorganisms13050992
Investigating Skin Microbial Community in Malignant Melanoma Lesions
Abstract
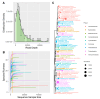
The skin microbiome is identified as one of the crucial factors in several pathological conditions, including its potential capacity in modulating cancer progression and response to treatment. A strong association of Bacilli and Betaproteobacteria classes and the Bacteroidetes phylum with melanoma is described in patients with cutaneous malignancies, while an imbalance of S. epidermidis and S. aureus is related to the progression of other skin cancers. In the present study, we characterized the microbial community in suspected lesions of 35 patients, classified, after histological analysis, as malignant melanoma lesions and benign non-melanoma lesions. Mirrored healthy skin were also included as negative control. No significant difference in alpha and beta diversity was observed when samples were categorized in four different groups (melanoma samples vs. contralateral healthy samples; melanoma samples vs. benign lesions; benign lesions vs. contralateral controls; melanoma controls vs. benign controls). The differential abundance analyses show that Corynebacterium urealyticum is more abundant in melanoma samples compared to their control, while Roseomonas gilardii is less abundant in melanoma. Staphylococcus massiliensis, Bacillus coagulans, Paracoccus yeei, Corynebacterium jeikeium, and Corynebacterium pyruviciproducens are present only in melanoma samples when compared with benign lesions.
Keywords: benign skin lesion; biomarker; melanoma; skin microbiota.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

References
-
- Kong H.H., Oh J., Deming C., Conlan S., Grice E.A., Beatson M.A., Nomicos E., Polley E.C., Komarow H.D., NISC Comparative Sequence Program et al. Temporal shifts in the skin microbiome associated with disease flares and treatment in children with atopic dermatitis. Genome Res. 2012;22:850–859. doi: 10.1101/gr.131029.111. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

