Transcriptomic Insights into the Degradation Mechanisms of Fomitopsis pinicola and Its Host Preference for Coniferous over Broadleaf Deadwood
- PMID: 40431179
- PMCID: PMC12113690
- DOI: 10.3390/microorganisms13051006
Transcriptomic Insights into the Degradation Mechanisms of Fomitopsis pinicola and Its Host Preference for Coniferous over Broadleaf Deadwood
Abstract
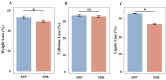
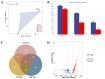
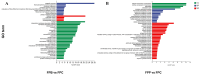
The degradation of deadwood is a vital ecological process for geochemical cycling and biodiversity conservation, with two main routes of fungal degradation: brown and white rot. Brown rot fungi cause severe destruction of wood cellulose and lead to brown and modified lignin residue. Fomitopsis pinicola is a typical brown rot fungus with a distinct host preference for coniferous trees. The mechanisms through which this fungus degrades coniferous and broadleaf wood remain poorly understood. Therefore, in this study, a 60-day cultivation experiment involving F. pinicola growing on deadwood strips of Pinus koraiensis and Betula platyphylla separately was performed. A comparative transcriptome analysis was carried out to explore the mechanisms underlying the differences in degradation, in terms of both physicochemical properties and transcriptomic data. The findings revealed that the host preference of F. pinicola resulted in the more efficient degradation of coniferous wood than broadleaf wood, accompanied by higher gene expression levels. GO enrichment analysis indicated that this preference was primarily associated with the hydrolytic enzyme family and processes related to the Fenton reaction, which is characteristic of brown rot fungi. Furthermore, the KEGG pathways showed that the DEGs were enriched in mainly included histidine metabolism, fatty acid degradation, and so on, indicating underlying carbohydrate and lipid metabolism processes. These results support P. pinicola's strong ability to degrade the deadwood lignin of P. koraiensis, reflecting its adaptive evolution in host selection and choice of different ecological niches.
Keywords: Fomitopsis pinicola; brown rot; comparative transcriptome analysis; degradation discrepancy mechanism.
Conflict of interest statement
The authors declare no conflicts of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of the data; in the writing of the manuscript; or in the decision to publish the results.
Figures

Similar articles
-
Polyporales Brown Rot Species Fomitopsis pinicola: Enzyme Activity Profiles, Oxalic Acid Production, and Fe3+-Reducing Metabolite Secretion.Appl Environ Microbiol. 2018 Apr 2;84(8):e02662-17. doi: 10.1128/AEM.02662-17. Print 2018 Apr 15. Appl Environ Microbiol. 2018. PMID: 29439983 Free PMC article.
-
Substrate-Specific Differential Gene Expression and RNA Editing in the Brown Rot Fungus Fomitopsis pinicola.Appl Environ Microbiol. 2018 Aug 1;84(16):e00991-18. doi: 10.1128/AEM.00991-18. Print 2018 Aug 15. Appl Environ Microbiol. 2018. Retracted and republished in: Appl Environ Microbiol. 2021 Jul 27;87(16):e0032921. doi: 10.1128/AEM.00329-21. Retraction in: Appl Environ Microbiol. 2021 Jul 27;87(16):e0033021. doi: 10.1128/AEM.00330-21. PMID: 29884757 Free PMC article. Retracted and republished. Retracted.
-
Retracted and Republished from: "Substrate-Specific Differential Gene Expression and RNA Editing in the Brown Rot Fungus Fomitopsis pinicola".Appl Environ Microbiol. 2021 Jul 27;87(16):e0032921. doi: 10.1128/AEM.00329-21. Epub 2021 Jul 27. Appl Environ Microbiol. 2021. PMID: 34313495 Free PMC article.
-
Decomposition of spruce wood and release of volatile organic compounds depend on decay type, fungal interactions and enzyme production patterns.FEMS Microbiol Ecol. 2019 Sep 1;95(9):fiz135. doi: 10.1093/femsec/fiz135. FEMS Microbiol Ecol. 2019. PMID: 31494677 Free PMC article.
-
Characterisation of Extracts and Anti-Cancer Activities of Fomitopsis pinicola.Nutrients. 2020 Feb 26;12(3):609. doi: 10.3390/nu12030609. Nutrients. 2020. PMID: 32110892 Free PMC article. Review.
References
-
- Harmon M., Franklin J., Swanson F., Sollins P., Gregory S., Lattin J., Anderson N. Ecology of coarse wooden debris in temperate ecosystems. Adv. Ecol. Res. 1986;15:133–302.
-
- Stokland J., Siitonen J., Jonsson B. Biodiversity in Dead Wood. Cambridge University Press; New York, NY, USA: 2012.
-
- Floudas D., Binder M., Riley R., Barry K., Blanchette R.A., Henrissat B., Martínez A.T., Otillar R., Spatafora J.W., Yadav J.S., et al. The paleozoic origin of enzymatic lignin decomposition reconstructed from 31 fungal genomes. Science. 2012;336:1715–1719. doi: 10.1126/science.1221748. - DOI - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

