Soil Microbial Adaptation and Biogeochemical Feedback in Degraded Alpine Meadows of the Qinghai-Tibetan Plateau
- PMID: 40431314
- PMCID: PMC12114374
- DOI: 10.3390/microorganisms13051142
Soil Microbial Adaptation and Biogeochemical Feedback in Degraded Alpine Meadows of the Qinghai-Tibetan Plateau
Abstract
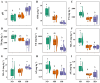
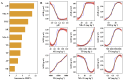
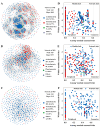
Alpine meadows on the Qinghai-Tibetan Plateau are experiencing rapid degradation due to climate change and anthropogenic disturbances, leading to severe ecological consequences. In this study, we investigated the response of soil microbial communities and their metabolic functions across a degradation gradient using metagenomic sequencing and comprehensive soil physicochemical analysis in the city of Lhasa, China. Results showed that soil pH increased with degradation, while most nutrients, including different forms of nitrogen, phosphorus, and potassium, declined. pH, ammonium nitrogen, and organic matter were identified as key factors driving degradation dynamics. Microbial community composition shifted markedly, with distinct biomarker taxa emerging at different degradation levels. Network analysis revealed a progressive loss of microbial connectivity, with Actinobacteria dominance increasing in heavily degraded soils, while cross-phylum interactions weakened. Functional analysis of biogeochemical cycling genes showed that carbon, nitrogen, and phosphorus cycling were all disrupted by degradation, but each exhibited unique response patterns. These findings will extend our understanding of microbial-mediated soil processes under degradation and provide a scientific foundation for ecosystem management, conservation, and targeted restoration strategies in alpine meadows.
Keywords: alpine meadow degradation; biogeochemical cycling; metagenomic sequencing; microorganism community; soil physicochemical properties.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures



Similar articles
-
Soil microbial diversity and composition response to degradation of the alpine meadow in the southeastern Qinghai-Tibet Plateau.Environ Sci Pollut Res Int. 2024 Apr;31(17):26076-26088. doi: 10.1007/s11356-024-32536-2. Epub 2024 Mar 16. Environ Sci Pollut Res Int. 2024. PMID: 38491240
-
Nutrients available in the soil regulate the changes of soil microbial community alongside degradation of alpine meadows in the northeast of the Qinghai-Tibet Plateau.Sci Total Environ. 2021 Oct 20;792:148363. doi: 10.1016/j.scitotenv.2021.148363. Epub 2021 Jun 9. Sci Total Environ. 2021. PMID: 34465051
-
Reduced soil multifunctionality and microbial network complexity in degraded and revegetated alpine meadows.J Environ Manage. 2023 Oct 1;343:118182. doi: 10.1016/j.jenvman.2023.118182. Epub 2023 May 22. J Environ Manage. 2023. PMID: 37224687
-
A functional metagenomics study of soil carbon and nitrogen degradation networks and limiting factors on the Tibetan plateau.Front Microbiol. 2023 May 5;14:1170806. doi: 10.3389/fmicb.2023.1170806. eCollection 2023. Front Microbiol. 2023. PMID: 37228377 Free PMC article.
-
[Vegetation and soil characteristics of degraded alpine meadows on the Qinghai-Tibet Pla-teau, China: A review].Ying Yong Sheng Tai Xue Bao. 2020 Jun;31(6):2109-2118. doi: 10.13287/j.1001-9332.202006.002. Ying Yong Sheng Tai Xue Bao. 2020. PMID: 34494765 Review. Chinese.
References
-
- Sun H., Liu J., Wu J., Hu H., Chen Q., Fang H., Tao K. Effects of alpine grassland degradation on soil microbial community structure and metabolic activity in the Qinghai-Tibet Plateau. Appl. Soil Ecol. 2024;200:105458. doi: 10.1016/j.apsoil.2024.105458. - DOI
-
- Li C., de Jong R., Schmid B., Wulf H., Schaepman M.E. Changes in grassland cover and in its spatial heterogeneity indicate degradation on the Qinghai-Tibetan Plateau. Ecol. Indic. 2020;119:106641. doi: 10.1016/j.ecolind.2020.106641. - DOI
-
- Qiao Y., Duan Z. Landscape and Ecosystem Diversity, Dynamics and Management in the Yellow River Source Zone. Springer; Cham, Switzerland: 2016. Understanding alpine meadow ecosystems; pp. 117–135.
Grants and funding
LinkOut - more resources
Full Text Sources

