A Novel Toolkit of SARS-CoV-2 Sub-Genomic Replicons for Efficient Antiviral Screening
- PMID: 40431609
- PMCID: PMC12115450
- DOI: 10.3390/v17050597
A Novel Toolkit of SARS-CoV-2 Sub-Genomic Replicons for Efficient Antiviral Screening
Abstract
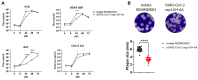
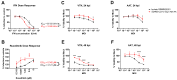
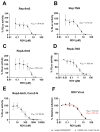
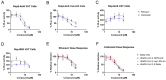
SARS-CoV-2 is classified as a containment level 3 (CL3) pathogen, limiting research access and antiviral testing. To address this, we developed a non-infectious viral surrogate system using reverse genetics to generate sub-genomic replicons. These replicons contained the nsp1 mutations K164A and H165A and had the spike, membrane, ORF6, and ORF7a coding sequences replaced with various reporter and selectable marker genes. Replicons based on the ancestral Wuhan Hu-1 strain and the Delta variant of concern were replication-competent in multiple cell lines, as assessed by Renilla luciferase activity, fluorescence, immunofluorescence staining, and single-molecule fluorescent in situ hybridization. Antiviral assays using transient replicon expression showed that remdesivir effectively inhibited both replicon and viral replication. Ritonavir and cobicistat inhibited Delta variant replicons similarly to wild-type virus but did not inhibit Wuhan Hu-1 replicon replication. To further investigate the impact of nsp1 mutations, we generated a recombinant SARS-CoV-2 virus carrying the K164A and H165A mutations. The virus exhibited attenuated replication across a range of mammalian cell lines, was restricted by the type I interferon response, and showed reduced cytopathic effects. These findings highlight the utility of sub-genomic replicons as reliable CL2-compatible surrogates for studying SARS-CoV-2 replication and drug activity mechanisms.
Keywords: RNA replicon; SARS-CoV-2; SARS-CoV-2 nsp1; antiviral.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures



References
-
- Gorbalenya A.E., Baker S.C., Baric R.S., De Groot R.J., Drosten C., Gulyaeva A.A., Haagmans B.L., Lauber C., Leontovich A.M., Neuman B.W., et al. Coronaviridae Study Group of the International Committee on Taxonomy of V. The species Severe acute respiratory syndrome-related coronavirus: Classifying 2019-nCoV and naming it SARS-CoV-2. Nat. Microbiol. 2020;5:536–544. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- MR/V027506/1; MR/R020566/1; MR/W005611/1/; MR/Y004205/1/MRC_/Medical Research Council/United Kingdom
- 794 204813/Z/16/Z; 200838/Z/16/Z/WT_/Wellcome Trust/United Kingdom
- 2018I2M-2-002/Chinese Academy of Medical Sciences, Innovation Fund for Medical Science
- 5F40120C00085/United States Food and Drug Administration
- MR/R020566/1/MRC_/Medical Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Miscellaneous

