Molecular Epidemiology of Travel-Associated and Locally Acquired Dengue Virus Infections in Catalonia, Spain, 2019
- PMID: 40431634
- PMCID: PMC12115671
- DOI: 10.3390/v17050621
Molecular Epidemiology of Travel-Associated and Locally Acquired Dengue Virus Infections in Catalonia, Spain, 2019
Abstract
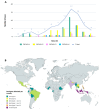
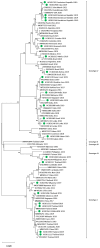
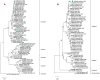
Dengue virus (DENV) is the most important arbovirus worldwide. In 2019, a significant increase in dengue cases was reported worldwide, resulting in a peak of imported cases in some European countries such as Spain. We aimed to describe travel-associated and locally acquired DENV strains detected in 2019 in the Catalonia region (northeastern Spain), a hotspot for dengue introduction in Europe. Through sequencing and phylogenetic analysis of the envelope gene, 75 imported viremic cases and two local strains were described. Autochthonous transmission events included an infection of a local mosquito with an imported dengue strain and a locally acquired human dengue infection from a locally infected mosquito. Overall, all four DENV serotypes and up to 10 different genotypes were detected. Phylogenetic analysis revealed transcontinental circulations associated with DENV-1 and DENV-2 and the presence of DENV-4 genotype I in Indonesia, where few cases had been previously described. A molecular study of the autochthonous events determined that local Ae. albopictus mosquitoes were infected by an African DENV-1 genotype V strain, while the locally acquired human case was caused by a DENV-3 genotype I of Asian origin. These findings underline the wide variability of imported strains and the high risk of DENV introduction into this territory, emphasizing the importance and usefulness of molecular characterization and phylogenetics for both local and global surveillance of the disease.
Keywords: Aedes albopictus; autochthonous transmission; dengue; molecular epidemiology; phylogenetic analysis; surveillance.
Conflict of interest statement
The authors declare no conflicts of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures




References
-
- World Health Organization Health Topics. Dengue and Severe Dengue. Updated 23 April 2024. [(accessed on 11 September 2024)]. Available online: https://www.who.int/news-room/fact-sheets/detail/dengue-and-severe-dengue.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

