Discovery and Genome Characterization of Three New Rhabdoviruses Infecting Passiflora spp. in Brazil
- PMID: 40431736
- PMCID: PMC12116104
- DOI: 10.3390/v17050725
Discovery and Genome Characterization of Three New Rhabdoviruses Infecting Passiflora spp. in Brazil
Abstract
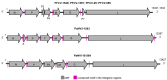
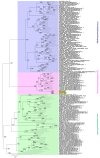
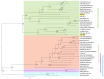
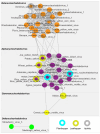
This study aimed to explore the RNA viruses affecting Passiflora species in Brazil. Our results enhance the understanding of the viruses that infect Passiflora plants by identifying and characterizing three previously unrecognized viruses: Passiflora cytorhabdovirus (PFCV), Passiflora nucleorhabdovirus 1 (PaNV1), and Passiflora nucleorhabdovirus 2 (PaNV2). These rhabdoviruses were identified through high-throughput sequencing and validated by reverse transcription-polymerase chain reaction (RT-PCR) in various Passiflora species. PFCV has a genome organization 3'-N-P-P3-P4-M-G-P7-L-5' and was classified as a novel member of the Gammacytorhabdovirus genus. A particularly noteworthy feature of PFCV is its glycoprotein, as the genomes of other gammarhabdoviruses do not contain this gene. PFCV has a high incidence across multiple locations and was identified in plants from Northeastern, Central, and Southeastern Brazil. PaNV1 with genome structure 3'-N-P-P3-M-G-L-5' and PaNV2 with genome organization 3'-N-X-P-Y-M-G-L-5' are new members of the Alphanucleorhabdovirus genus and have a more restricted occurrence. Importantly, all three viruses were found in mixed infections alongside at least one other virus. In situ observations confirmed mixed infections, with PaNV2 particles co-located in tissues with a potyvirus and a carlavirus. Phylogenetic and glycoprotein sequence similarity network analysis provided insights into their evolutionary placement and potential vector associations. These findings expand the known diversity of rhabdoviruses in Passiflora and contribute to the understanding of their evolution and epidemiology.
Keywords: alphanucleorhabdovirus; gammacytorhabdovirus; high-throughput sequencing; microscopy; plant rhabdovirus; virus particles.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures


Similar articles
-
Genomes of Alphanucleorhabdovirus Physostegiae Isolates from Two Different Cultivar Groups of Solanum melongena.Viruses. 2024 Sep 28;16(10):1538. doi: 10.3390/v16101538. Viruses. 2024. PMID: 39459872 Free PMC article.
-
Detection and In Vivo Validation of Dichorhavirus e-Probes in Meta-Transcriptomic Data via Microbe Finder (MiFi®) Discovers a Novel Host and a Possible New Strain of Orchid Fleck Virus.Viruses. 2025 Mar 19;17(3):441. doi: 10.3390/v17030441. Viruses. 2025. PMID: 40143368 Free PMC article.
-
Merida virus, a putative novel rhabdovirus discovered in Culex and Ochlerotatus spp. mosquitoes in the Yucatan Peninsula of Mexico.J Gen Virol. 2016 Apr;97(4):977-987. doi: 10.1099/jgv.0.000424. Epub 2016 Feb 11. J Gen Virol. 2016. PMID: 26868915 Free PMC article.
-
Signs and symptoms to determine if a patient presenting in primary care or hospital outpatient settings has COVID-19.Cochrane Database Syst Rev. 2022 May 20;5(5):CD013665. doi: 10.1002/14651858.CD013665.pub3. Cochrane Database Syst Rev. 2022. PMID: 35593186 Free PMC article.
-
Systemic pharmacological treatments for chronic plaque psoriasis: a network meta-analysis.Cochrane Database Syst Rev. 2021 Apr 19;4(4):CD011535. doi: 10.1002/14651858.CD011535.pub4. Cochrane Database Syst Rev. 2021. Update in: Cochrane Database Syst Rev. 2022 May 23;5:CD011535. doi: 10.1002/14651858.CD011535.pub5. PMID: 33871055 Free PMC article. Updated.
References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

