Transcriptomic insights into Mycobacterium orygis infection-associated pulmonary granulomas reveal multicellular immune networks and tuberculosis biomarkers in cattle
- PMID: 40432328
- PMCID: PMC12120866
- DOI: 10.1080/01652176.2025.2509503
Transcriptomic insights into Mycobacterium orygis infection-associated pulmonary granulomas reveal multicellular immune networks and tuberculosis biomarkers in cattle
Abstract
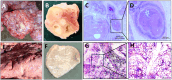
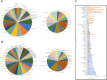
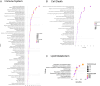
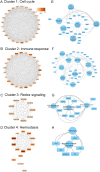
Mycobacterium orygis, a member of the Mycobacterium tuberculosis complex (MTBC), has emerged as a significant contributor to tuberculosis (TB) in cattle, wildlife, and humans. However, understanding about its pathogenesis and severity is limited, compounded by the lack of reliable TB biomarkers in cattle. This study delves into the comparative pathology and transcriptomic landscape of pulmonary granulomas in cattle naturally infected with M. orygis, using high-throughput RNA sequencing. Histopathological analysis revealed extensive, multistage granulomatous, necrotic, and cavitary lesions, indicative of severe lung pathology induced by M. orygis. Transcriptomic profiling highlighted numerous differentially expressed genes and dysregulated pathways related to immune response modulation and extracellular matrix remodelling. Additionally, cell type enrichment analysis provided insights into the multicellularity of the granulomatous niche, emphasising complex cell-cell interactions within TB granulomas. Via comparative transcriptomics leveraging publicly available bovine and human TB omics datasets, 14 key immunomodulators (SOD2, IL1α/β, IL15, IL18, CCL2/MCP-1, CCL3/MIP-1α, CCL4/MIP-1β, CCL8/MCP-2, CCL20/MIP-3α, CXCL2/MIP-2, CXCL10/IP-10, CXCL11, and IFN-γ) were identified as potential biomarkers for active TB in cattle. These findings significantly advance our understanding of M. orygis pathogenesis in bovine TB and highlight potential targets for the development of diagnostic tools for managing and controlling the disease.
Keywords: Mycobacterium orygis; RNA sequencing; Tuberculosis; biomarkers; bovine tuberculosis; granuloma; transcriptome; zoonotic TB.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures



References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous
