Long Noncoding RNA NONHSAT233728.1 Promotes ROS Accumulation and Granulosa Cell Apoptosis by Regulating the MAPK/ERK1/2 Signaling Pathway
- PMID: 40432414
- PMCID: PMC12117357
- DOI: 10.1096/fj.202500964R
Long Noncoding RNA NONHSAT233728.1 Promotes ROS Accumulation and Granulosa Cell Apoptosis by Regulating the MAPK/ERK1/2 Signaling Pathway
Abstract
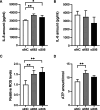
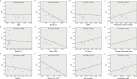
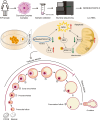
Polycystic ovary syndrome (PCOS) is one of the most prevalent endocrine disorders in women of reproductive age. However, the underlying molecular mechanism remains unclear. In this study, we employed RNA sequencing analysis to identify differentially expressed protein-coding genes and long noncoding RNA (lncRNA) expression profiles in granulosa cells from women with and without PCOS. It was established that the level of NONHSAT233728.1 was diminished in women with PCOS. The present study demonstrated the role of NONHSAT233728.1 in granulosa cells from patients with PCOS and further investigated the potential mechanism of NONHSAT233728.1 in the KGN cell line. Additionally, the knockdown of NONHSAT233728.1 has been observed to promote cell apoptosis, inhibit cell proliferation, promote mitochondrial dysfunction, and inflammation. Western blot analyses confirmed that phospho-extracellular regulated protein kinases (ERK)1/2 were decreased following lnc-NONHSAT233728.1 knockdown. Consequently, we propose that ROS accumulation activates the endogenous mitochondrial apoptosis pathway, leading to granulosa cell apoptosis via the MEK/ERK1/2 pathway, which contributes to follicular atresia. We observed a negative correlation between NONHSAT233728.1 and both LH levels and the LH/FSH ratio. These findings indicate that lncRNA NONHSAT233728.1 is linked to the pathogenesis of PCOS and offer new insights into its underlying mechanisms.
Keywords: apoptosis; granulosa cell; lncRNA; polycystic ovary syndrome; reactive oxygen species.
© 2025 The Author(s). The FASEB Journal published by Wiley Periodicals LLC on behalf of Federation of American Societies for Experimental Biology.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures



Similar articles
-
Follicular fluid-derived exosomal LncRNA LIPE-AS1 modulates steroid metabolism and survival of granulosa cells leading to oocyte maturation arrest in polycystic ovary syndrome.J Assist Reprod Genet. 2024 May;41(5):1387-1401. doi: 10.1007/s10815-024-03092-y. Epub 2024 Apr 24. J Assist Reprod Genet. 2024. PMID: 38656738 Free PMC article.
-
Functional microarray analysis of differentially expressed genes in granulosa cells from women with polycystic ovary syndrome related to MAPK/ERK signaling.Sci Rep. 2015 Oct 13;5:14994. doi: 10.1038/srep14994. Sci Rep. 2015. PMID: 26459919 Free PMC article.
-
Down-regulation of long non-coding RNA MALAT1 inhibits granulosa cell proliferation in endometriosis by up-regulating P21 via activation of the ERK/MAPK pathway.Mol Hum Reprod. 2019 Jan 1;25(1):17-29. doi: 10.1093/molehr/gay045. Mol Hum Reprod. 2019. PMID: 30371869
-
Long noncoding RNAs as a piece of polycystic ovary syndrome puzzle.Mol Biol Rep. 2021 Apr;48(4):3845-3851. doi: 10.1007/s11033-021-06196-1. Epub 2021 May 15. Mol Biol Rep. 2021. PMID: 33993404 Review.
-
Mechanisms of Granulosa Cell Programmed Cell Death and Follicular Atresia in Polycystic Ovary Syndrome.Physiol Res. 2025 Mar 21;74(1):31-40. doi: 10.33549/physiolres.935485. Physiol Res. 2025. PMID: 40116548 Free PMC article. Review.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

