Epigenetic mechanisms involved in hepatocellular carcinoma development and progression
- PMID: 40432834
- PMCID: PMC12107448
- DOI: 10.1136/egastro-2025-100186
Epigenetic mechanisms involved in hepatocellular carcinoma development and progression
Abstract
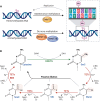
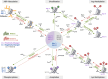
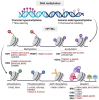
Hepatocellular carcinoma (HCC) typically develops in the context of chronic liver disease, where prolonged hepatocyte exposure to inflammation drives the synergistic accumulation of genetic and epigenetic alterations. Epigenetic regulation encompasses multiple mechanisms that govern the transcription machinery accessibility to DNA. This process is regulated by the addition and removal of covalent marks on chromatin, which can either affect DNA-histone interactions or serve as scaffolds for other proteins, among other mechanisms. Recent research has revealed that epigenetic alterations can disrupt chromatin homeostasis, redirecting transcriptional regulation to favour cancer-promoting states. Consequently, these alterations play a pivotal role in the acquisition of cancer hallmarks and provide insights into several biological processes involved in hepatocarcinogenesis. This review highlights the key epigenetic mechanisms underlying the development, progression and dissemination of HCC, with a particular focus on DNA methylation and histone post-translational modifications. This knowledge is relevant for guiding the development of innovative therapeutic approaches based on epigenetic modulators.
Keywords: Epigenetics; Gastrointestinal Cancer; Hepatocarcinogenesis; Hepatocellular Carcinoma.
Copyright © Author(s) (or their employer(s)) 2025. Re-use permitted under CC BY-NC. No commercial re-use. See rights and permissions. Published by BMJ Group.
Conflict of interest statement
BB is an employee of HZ4 Liver Inc. / Spectrum, Dover, Delaware, USA.
Figures
References
Publication types
LinkOut - more resources
Full Text Sources
