Bioinformatics Analysis of Lactylation-related Biomarkers and Potential Pathogenesis Mechanisms in Age-related Macular Degeneration
- PMID: 40433417
- PMCID: PMC12105335
- DOI: 10.2174/0113892029291661241114055924
Bioinformatics Analysis of Lactylation-related Biomarkers and Potential Pathogenesis Mechanisms in Age-related Macular Degeneration
Abstract
Background: Lactylation is increasingly recognized to play a crucial role in human health and diseases. However, its involvement in age-related macular degeneration (AMD) remains largely unclear.
Objectives: The aim of this study was to identify and characterize the pivotal lactylation-related genes and explore their underlying mechanism in AMD.
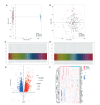
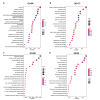
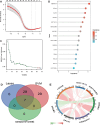
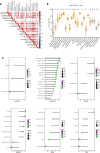
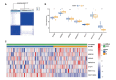
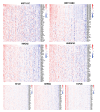
Methods: Gene expression profiles of AMD patients and control individuals were obtained and integrated from the GSE29801 and GSE50195 datasets. Differentially expressed genes (DEGs) were screened and intersected with lactylation-related genes for lactylation-related DEGs. Machine learning algorithms were used to identify hub genes associated with AMD. Subsequently, the selected hub genes were subject to correlation analysis, and reverse transcription quantitative real-time PCR (RT-qPCR) was used to detect the expression of hub genes in AMD patients and healthy control individuals.
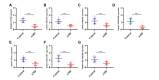
Results: A total of 68 lactylation-related DEGs in AMD were identified, and seven genes, including HMGN2, TOP2B, HNRNPH1, SF3A1, SRRM2, HIST1H1C, and HIST1H2BD were selected as key genes. RT-qPCR analysis validated that all 7 key genes were down-regulated in AMD patients.
Conclusion: We identified seven lactylation-related key genes potentially associated with the progression of AMD, which might deepen our understanding of the underlying mechanisms involved in AMD and provide clues for the targeted therapy.
Keywords: Age-related macular degeneration; bioinformatics; hub genes; immune infiltration; lactylation; machine learning.
© 2025 The Author(s). Published by Bentham Science Publisher.
Conflict of interest statement
The authors declare no conflict of interest, financial or otherwise.
Figures




Similar articles
-
Identification of Biomarkers Co-Associated with Lactylation and Acetylation in Systemic Lupus Erythematosus.Biomedicines. 2025 May 22;13(6):1274. doi: 10.3390/biomedicines13061274. Biomedicines. 2025. PMID: 40563993 Free PMC article.
-
Ferroptosis-related hub genes and immune cell dynamics as diagnostic biomarkers in age-related macular degeneration.Eur J Med Res. 2025 Aug 20;30(1):777. doi: 10.1186/s40001-025-03044-x. Eur J Med Res. 2025. PMID: 40830492 Free PMC article.
-
Deciphering Shared Gene Signatures and Immune Infiltration Characteristics Between Gestational Diabetes Mellitus and Preeclampsia by Integrated Bioinformatics Analysis and Machine Learning.Reprod Sci. 2025 Jun;32(6):1886-1904. doi: 10.1007/s43032-025-01847-1. Epub 2025 May 15. Reprod Sci. 2025. PMID: 40374866
-
Surgery for cataracts in people with age-related macular degeneration.Cochrane Database Syst Rev. 2017 Feb 16;2(2):CD006757. doi: 10.1002/14651858.CD006757.pub4. Cochrane Database Syst Rev. 2017. PMID: 28206671 Free PMC article.
-
Blue-light filtering intraocular lenses (IOLs) for protecting macular health.Cochrane Database Syst Rev. 2018 May 22;5(5):CD011977. doi: 10.1002/14651858.CD011977.pub2. Cochrane Database Syst Rev. 2018. PMID: 29786830 Free PMC article.
References
-
- Lee Q., Chan W.C., Qu X., Sun Y., Abdelkarim H., Le J., Saqib U., Sun M.Y., Kruse K., Banerjee A., Hitchinson B., Geyer M., Huang F., Guaiquil V., Mutso A.A., Sanders M., Rosenblatt M.I., Maienschein-Cline M., Lawrence M.S., Gaponenko V., Malik A.B., Komarova Y.A. End binding-3 inhibitor activates regenerative program in age-related macular degeneration. Cell Rep. Med. 2023;4(10):101223. doi: 10.1016/j.xcrm.2023.101223. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous
