Protein Secondary Structure Patterns in Short-Range Cross-Link Atlas
- PMID: 40433759
- PMCID: PMC12304803
- DOI: 10.1002/anie.202507348
Protein Secondary Structure Patterns in Short-Range Cross-Link Atlas
Abstract
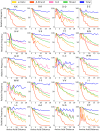
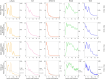
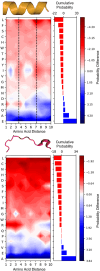
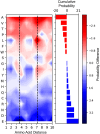
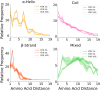
Cross-linking mass spectrometry (XL-MS) has become a powerful tool in structural biology for investigating protein structure, dynamics, and interactomics. However, short-range cross-links, defined as those connecting residues fewer than 20 positions apart, have traditionally been considered less informative and largely overlooked, leaving significant data unexplored in a systematic manner. Here, we present a system-wide analysis of short-range cross-links, demonstrating their intrinsic correlation with protein secondary structure. We introduce the X-SPAN (Cross-link Structural Pattern Analyzer) software, which integrates publicly available XL-MS datasets from system-wide experiments with AlphaFold-predicted protein structures. Our analysis reveals distinct cross-linking patterns that reflect the spatial constraints imposed by secondary structural elements. Specifically, α-helices exhibit periodic cross-linking patterns consistent with their characteristic helical pitch, whereas coils and β-strands display nearly monotonic distributions. A context-dependent protein grammar reinforces short-range cross-link specificity. Short-range cross-links can enhance the statistical inference of secondary structures within integrative modeling workflows. Additionally, our work establishes a framework for benchmarking AlphaFold's local prediction accuracy and provides novel quality control criteria for XL-MS experiments. We anticipate that X-SPAN and our short-range cross-link database will serve as a valuable resource for exploring local secondary structure rearrangements and their potential roles in protein function and allosteric regulation.
Keywords: Chemical proteomics; Cross‐linking; Mass spectrometry; Structural biology; Structural proteomics.
© 2025 The Author(s). Angewandte Chemie International Edition published by Wiley‐VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
MeSH terms
Substances
Grants and funding
- PRIN 2022/Italian Ministry of University and Research (MUR)
- Project 20225HNCZK/Italian Ministry of University and Research (MUR)
- Master CUP G53D23002450006/Italian Ministry of University and Research (MUR)
- CUP E53D23007110006/Italian Ministry of University and Research (MUR)
- PRIN 2022 PNRR/European Union NextGeneration EU
- Project P20224WAME/European Union NextGeneration EU
- CUP E53D23021440001/European Union NextGeneration EU
- ECS00000041/European Union NextGeneration EU under the Italian Ministry of University and Research (MUR) National Innovation Ecosystem
- VITALITY/European Union NextGeneration EU under the Italian Ministry of University and Research (MUR) National Innovation Ecosystem
- CUP E13C22001060006/European Union NextGeneration EU under the Italian Ministry of University and Research (MUR) National Innovation Ecosystem
LinkOut - more resources
Full Text Sources

