Contrasting interferon-mediated antiviral responses in human lung adenocarcinoma cells
- PMID: 40434103
- PMCID: PMC12172473
- DOI: 10.1128/jvi.00469-25
Contrasting interferon-mediated antiviral responses in human lung adenocarcinoma cells
Abstract
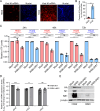
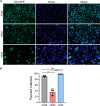
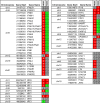
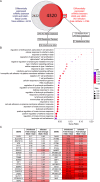
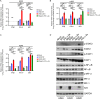
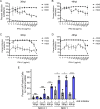
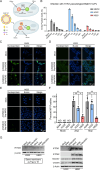
Lung cancers develop from lung epithelial cells after a series of genetic and epigenetic changes, and these cells are major sites of influenza virus infection. Thus, we explored how changes found in patient-derived lung cancer cell lines impacted influenza virus replication and identified two lines with opposite responses to influenza A viral infection. We show that the NCI-H820 lung adenocarcinoma (LUAD) is resistant to influenza A virus and VSV infection, while LUAD line NCI-H322 is highly susceptible to infection by both viruses. H322 cells have a homozygous deletion in a region of chromosome 9 encoding IFNαgenes, IFNβ1, IFNω1, and IFNε genes, leading to downregulation of immune response and high infection rates. In contrast, the resistant H820 cell line has three copies of these same interferon genes and shows increased expression of interferon-regulated genes. We found that the resistance of H820 cells to influenza infection is likely linked to impaired viral entry-due to high basal levels of interferon-induced proteins known to inhibit endocytosis (IFITM1/2/3, NCOA7, and CH25H)-and to increased expression of mRNAs that encode other antiviral factors. In contrast, H322 cells show the absence or low levels of interferon-regulated genes involved in the inhibition of viral entry. These results suggest that the opposite phenotypes on viral entry of H322 and H820 cells may be at least in part associated with impaired or enhanced interferon response, respectively. Since most lung cancer patients have genomic characterization of their tumors, individualized differences in interferon responses may have therapeutic and patient management implications.
Importance: Lung cancers develop from genetic and epigenetic changes that can dramatically influence patients' susceptibility to viral infection and replication. This study evaluates the responses to influenza virus infection of two patient-derived lung cancer cell lines. Interestingly, the cell lines investigated are of the same cancer type, lung adenocarcinomas, yet one cell line is highly susceptible, while the other cell line is highly resistant to viral infection. This is in part due to contrasting genetic alterations that lead to changes in the interferon response pathways, which differentially impact viral entry. Thus, identifying these risk factors can inform the prognosis of patients infected with influenza virus and guide their personalized treatment plans.
Keywords: influenza; interferons; lung cancer; viral entry.
Conflict of interest statement
The A.G.-S. laboratory has received research support from GSK, Pfizer, Senhwa Biosciences, Kenall Manufacturing, Blade Therapeutics, Avimex, Johnson and Johnson, Dynavax, 7Hills Pharma, Pharmamar, ImmunityBio, Accurius, Nanocomposix, Hexamer, N-fold LLC, Model Medicines, Atea Pharma, Applied Biological Laboratories, and Merck, outside of the reported work. A.G.-S. has consulting agreements for the following companies involving cash and/or stock: Castlevax, Amovir, Vivaldi Biosciences, Contrafect, 7Hills Pharma, Avimex, Pagoda, Accurius, Esperovax, Applied Biological Laboratories, Pharmamar, CureLab Oncology, CureLab Veterinary, Synairgen, Paratus, Pfizer, and Prosetta, outside of the reported work. A.G.-S. has been an invited speaker in meeting events organized by Seqirus, Janssen, Abbott, Astrazeneca, and Novavax. A.G.-S. is the inventor on patents and patent applications on the use of antivirals and vaccines for the treatment and prevention of virus infections and cancer, owned by the Icahn School of Medicine at Mount Sinai, New York, outside of the reported work.
Figures
Similar articles
-
METTL3 depletion blocks vesicular stomatitis virus replication in pancreatic cancer cells through the establishment of an intrinsic antiviral state.J Virol. 2025 May 20;99(5):e0228424. doi: 10.1128/jvi.02284-24. Epub 2025 Apr 11. J Virol. 2025. PMID: 40214229 Free PMC article.
-
CRISPR editing of candidate host factors that impact influenza A virus infection.Microbiol Spectr. 2025 Mar 4;13(3):e0262724. doi: 10.1128/spectrum.02627-24. Epub 2025 Jan 31. Microbiol Spectr. 2025. PMID: 39887213 Free PMC article.
-
Ineffectual immunity in a resurrected mouse model of persistent viremia.J Virol. 2025 Jun 17;99(6):e0024825. doi: 10.1128/jvi.00248-25. Epub 2025 May 8. J Virol. 2025. PMID: 40338081 Free PMC article.
-
Assessing the comparative effects of interventions in COPD: a tutorial on network meta-analysis for clinicians.Respir Res. 2024 Dec 21;25(1):438. doi: 10.1186/s12931-024-03056-x. Respir Res. 2024. PMID: 39709425 Free PMC article. Review.
-
Mitigation and Management of Common Toxicities Associated with the Administration of CAR-T Therapies in Oncology Patients.Drug Saf. 2025 Jul;48(7):719-737. doi: 10.1007/s40264-025-01538-5. Epub 2025 Mar 19. Drug Saf. 2025. PMID: 40108072 Free PMC article. Review.
References
-
- Chatterjee S, Crozet L, Damotte D, Iribarren K, Schramm C, Alifano M, Lupo A, Cherfils-Vicini J, Goc J, Katsahian S, Younes M, Dieu-Nosjean MC, Fridman WH, Sautès-Fridman C, Cremer I. 2014. TLR7 promotes tumor progression, chemotherapy resistance, and poor clinical outcomes in non-small cell lung cancer. Cancer Res 74:5008–5018. doi: 10.1158/0008-5472.CAN-13-2698 - DOI - PubMed
MeSH terms
Substances
Grants and funding
- U54 CA260560/CA/NCI NIH HHS/United States
- R01 HL089966/HL/NHLBI NIH HHS/United States
- P50 CA070907/NH/NIH HHS/United States
- P30 CA142543-06/NH/NIH HHS/United States
- P30 CA142543/CA/NCI NIH HHS/United States
- 1S10OD018005-01/NH/NIH HHS/United States
- P50 CA070907/CA/NCI NIH HHS/United States
- 75N93021C00014/NH/NIH HHS/United States
- S10 OD018005/OD/NIH HHS/United States
- R01 AI154635/NH/NIH HHS/United States
- U54 CA260560/NH/NIH HHS/United States
- R01 AI154635/AI/NIAID NIH HHS/United States
- U19 AI135972/AI/NIAID NIH HHS/United States
- 75N93021C00014/AI/NIAID NIH HHS/United States
- HL089966/NH/NIH HHS/United States
- U19AI135972/NH/NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical

