Neuronal potassium channel activity triggers initiation of mRNA translation through binding of translation regulators
- PMID: 40435242
- PMCID: PMC12118559
- DOI: 10.1126/sciadv.adv3140
Neuronal potassium channel activity triggers initiation of mRNA translation through binding of translation regulators
Abstract
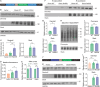
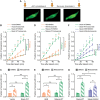
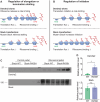
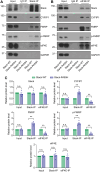
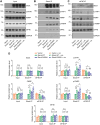
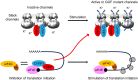
Neuronal activity stimulates mRNA translation crucial for learning and development, but the mechanism linking translation to neuronal activity is not understood. In humans, learning and memory are severely disrupted by mutations in the potassium channel Slack (KCNT1, Slo2.2). We find that pharmacological stimulation of this channel and a constitutively active Slack mutation stimulate mRNA translation of a reporter for β-actin mRNA in cell lines and increases the synthesis of β-actin in the neurites of cortical neurons. Moreover, channel activation promotes the binding of two key mRNA translation regulators, FMRP (fragile X mental retardation protein) and CYFIP1 (cytoplasmic FMR1-interacting protein 1), to the channel itself, releasing both from eIF4E (eukaryotic initiation factor 4E), where they normally inhibit initiation of translation. This interaction provides a molecular mechanism for Slack activity-dependent regulation of translation and suggests that the effects of Slack mutations on this process may explain the severe intellectual disabilities associated with these mutations.
Figures


Update of
-
Neuronal potassium channel activity triggers initiation of mRNA translation through binding of translation regulators.bioRxiv [Preprint]. 2024 Feb 7:2024.02.07.579306. doi: 10.1101/2024.02.07.579306. bioRxiv. 2024. Update in: Sci Adv. 2025 May 30;11(22):eadv3140. doi: 10.1126/sciadv.adv3140. PMID: 38370631 Free PMC article. Updated. Preprint.
References
-
- Bhattacharjee A., Kaczmarek L. K., For K+ channels, Na+ is the new Ca2+. Trends Neurosci. 28, 422–428 (2005). - PubMed
-
- Joiner W. J., Tang M. D., Wang L. Y., Dworetzky S. I., Boissard C. G., Gan L., Gribkoff V. K., Kaczmarek L. K., Formation of intermediate-conductance calcium-activated potassium channels by interaction of Slack and Slo subunits. Nat. Neurosci. 1, 462–469 (1998). - PubMed
-
- Wu J., El-Hassar L., Datta D., Thomas M., Zhang Y., Jenkins D. P., DeLuca N. J., Chatterjee M., Gribkoff V. K., Arnsten A. F. T., Kaczmarek L. K., Interaction between HCN and slack channels regulates mPFC pyramidal cell excitability in working memory circuits. Mol. Neurobiol. 61, 2430–2445 (2024). - PubMed
-
- Bhattacharjee A., von Hehn C. A., Mei X., Kaczmarek L. K., Localization of the Na+−activated K+ channel Slick in the rat central nervous system. J. Comp. Neurol. 484, 80–92 (2005). - PubMed
-
- Bhattacharjee A., Gan L., Kaczmarek L. K., Localization of the Slack potassium channel in the rat central nervous system. J. Comp. Neurol. 454, 241–254 (2002). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

