PIEZO1-mediated calcium signaling reinforces mechanical properties of hair follicle stem cells to promote quiescence
- PMID: 40435254
- PMCID: PMC12118625
- DOI: 10.1126/sciadv.adt2771
PIEZO1-mediated calcium signaling reinforces mechanical properties of hair follicle stem cells to promote quiescence
Abstract
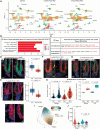
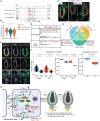
The mechanisms by which epithelial stem cells (SCs) sense mechanical cues within their niche and convert the information into biochemical signals to govern their function are not well understood. Here, we show that hair follicle SCs (HF-SCs) sense mechanical forces through cell adhesion and maintain quiescence in a PIEZO1-dependent mechanism. PIEZO1 interacts with E-cadherin in HF-SCs, and mechanical pulling of E-cadherin with a force of ~20 pN triggers PIEZO1-dependent, localized calcium flickers. Deletion of Piezo1 leads to reduced cumulative calcium influx and compromises quiescence. Single-cell genomic analyses identify a transcriptional network involving AP1 and NFATC1, which functions downstream of PIEZO1 and regulates the expression of extracellular matrix, cell adhesion, and actin cytoskeleton genes to reinforce the unique mechanical property of HF-SCs. These findings establish the force threshold necessary for PIEZO1 activation and reveal PIEZO1-dependent calcium influx as a key mechanism for sensing mechanical cues in the niche and regulating HF-SC activity.
Figures






References
-
- Engler A. J., Sen S., Sweeney H. L., Discher D. E., Matrix elasticity directs stem cell lineage specification. Cell 126, 677–689 (2006). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

