Adaptation of DNA to Protein Binding Revealed by Spectroscopy and Molecular Simulation
- PMID: 40436634
- PMCID: PMC12169650
- DOI: 10.1021/acs.jpcb.5c00189
Adaptation of DNA to Protein Binding Revealed by Spectroscopy and Molecular Simulation
Abstract
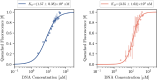
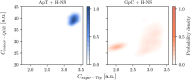
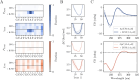
DNA demonstrates remarkable structural diversity, transitioning between conformations such as B-DNA and A-DNA under specific environmental or protein-binding conditions. These transitions are relevant for mediating cellular processes such as gene regulation, DNA organization, and stress response. In bacteria, the histone-like nucleoid structuring protein (H-NS) exemplifies the interaction between sequence-dependent DNA conformational adaptability and protein-mediated regulatory mechanisms. Despite evidence for the strong affinity of H-NS for AT-rich DNA, the specific molecular and structural interactions driving this recognition remain largely unclear. Combining fluorescence spectroscopy, circular dichroism (CD), molecular dynamics (MD) simulations, and enhanced sampling techniques, we show that H-NS exhibits a 10-fold higher affinity for ApT repeats compared to that of GpC repeats. Interestingly, selective binding of H-NS to AT-rich DNA causes a structural adaptation in the DNA, including increased bending flexibility, minor groove widening, and localized A-like DNA features, while GC-rich DNA remains closer to the canonical B-form. Our approach yielded detailed insights into how H-NS exploits the intrinsic conformational plasticity of DNA to achieve sequence-dependent binding. More broadly, this work illustrates how DNA-binding proteins can harness the structural adaptability of the DNA double helix, which may modulate regulatory outcomes, and provides insight into how the intrinsic properties of DNA shape protein-DNA interactions in diverse biological systems.
Figures

References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

