Tracing the spatial origins and spread of SARS-CoV-2 Omicron lineages in South Africa
- PMID: 40436832
- PMCID: PMC12120024
- DOI: 10.1038/s41467-025-60081-0
Tracing the spatial origins and spread of SARS-CoV-2 Omicron lineages in South Africa
Abstract
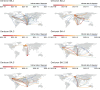
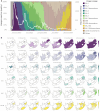
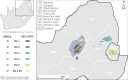
Since November 2021, five genetically distinct SARS-CoV-2 Omicron lineages (BA.1-BA.5) are believed to have emerged in southern Africa, with four (BA.1, BA.2, BA.4, and BA.5) spreading globally and collectively dominating SARS-CoV-2 diversity. In 2023, BA.2.86, a highly divergent BA.2 lineage that rose to prominence worldwide, was first detected in Israel and Denmark, but the subsequent diversity of South African sequences suggests it too emerged in the region. Using Bayesian phylogeographic inference, we reconstruct the origins and dispersal patterns of BA.1-BA.5 and BA.2.86. Our findings suggest that Gauteng province in South Africa likely played a key role in the emergence and/or amplification of multiple Omicron lineages, though regions with limited sampling may have also contributed. The challenge of precisely tracing these origins highlights the need for broader genomic surveillance across the region to strengthen early detection, track viral evolution, and improve preparedness for future threats.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: N.W. has received grant funding from the US CDC, Gates Foundation and Sanofi. The remaining authors declare no competing interests.
Figures

References
-
- Tegally, H. et al. Detection of a SARS-CoV-2 variant of concern in South Africa. Nature592, 438–443 (2021). - PubMed
-
- Stats, S. A. Mid-Year Population Estimates. https://www.statssa.gov.za/?page_id=1854 (2022).
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

