ACE2 from Pipistrellus abramus bats is a receptor for HKU5 coronaviruses
- PMID: 40436893
- PMCID: PMC12119886
- DOI: 10.1038/s41467-025-60286-3
ACE2 from Pipistrellus abramus bats is a receptor for HKU5 coronaviruses
Abstract
The merbecovirus subgenus of coronaviruses includes Middle East Respiratory Syndrome Coronavirus (MERS-CoV), a zoonotic pathogen transmitted from dromedary camels to humans that causes severe respiratory disease. Viral discovery efforts uncover hundreds of merbecoviruses in different species across multiple continents, but few are studied under laboratory conditions, leaving basic questions regarding their human threat potential unresolved. Viral entry into host cells is a critical step for transmission between hosts. Here, we develop and apply a scalable approach to assesses novel merbecovirus cell entry across the entire merbecovirus subgenus. Merbecoviruses are sorted into clades based on the receptor-binding domain of the spike glycoprotein. Receptor tropism is clade-specific, with the clade including MERS-CoV using DPP4 and multiple clades using ACE2, including HKU5 bat coronaviruses. Mutational analysis identifies possible structural limitations to HKU5 adaptability and a cryo-EM structure of the HKU5-20s spike trimer reveals only 'down' RBDs.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: RSB is a member of advisory boards for VaxArt, Takeda and Invivyd, and has collaborative projects with Gilead, J&J, and Hillevax, focused on unrelated projects. The remaining authors compare no competing interests.
Figures


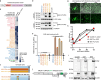
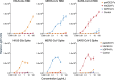



Update of
-
ACE2 from Pipistrellus abramus bats is a receptor for HKU5 coronaviruses.bioRxiv [Preprint]. 2024 Aug 16:2024.03.13.584892. doi: 10.1101/2024.03.13.584892. bioRxiv. 2024. Update in: Nat Commun. 2025 May 28;16(1):4932. doi: 10.1038/s41467-025-60286-3. PMID: 38559009 Free PMC article. Updated. Preprint.
Similar articles
-
ACE2 from Pipistrellus abramus bats is a receptor for HKU5 coronaviruses.bioRxiv [Preprint]. 2024 Aug 16:2024.03.13.584892. doi: 10.1101/2024.03.13.584892. bioRxiv. 2024. Update in: Nat Commun. 2025 May 28;16(1):4932. doi: 10.1038/s41467-025-60286-3. PMID: 38559009 Free PMC article. Updated. Preprint.
-
Bat-infecting merbecovirus HKU5-CoV lineage 2 can use human ACE2 as a cell entry receptor.Cell. 2025 Mar 20;188(6):1729-1742.e16. doi: 10.1016/j.cell.2025.01.042. Epub 2025 Feb 18. Cell. 2025. PMID: 39970913
-
Molecular basis of convergent evolution of ACE2 receptor utilization among HKU5 coronaviruses.Cell. 2025 Mar 20;188(6):1711-1728.e21. doi: 10.1016/j.cell.2024.12.032. Epub 2025 Feb 7. Cell. 2025. PMID: 39922192 Free PMC article.
-
The lethal internal face of the coronaviruses: Kidney tropism of the SARS, MERS, and COVID19 viruses.IUBMB Life. 2021 Aug;73(8):1005-1015. doi: 10.1002/iub.2516. Epub 2021 Jun 30. IUBMB Life. 2021. PMID: 34118117 Free PMC article. Review.
-
Unraveling the Epidemiology, Geographical Distribution, and Genomic Evolution of Potentially Lethal Coronaviruses (SARS, MERS, and SARS CoV-2).Front Cell Infect Microbiol. 2020 Aug 27;10:499. doi: 10.3389/fcimb.2020.00499. eCollection 2020. Front Cell Infect Microbiol. 2020. PMID: 32974224 Free PMC article. Review.
Cited by
-
ACE2 utilization of HKU25 clade MERS-related coronaviruses with broad geographic distribution.bioRxiv [Preprint]. 2025 Feb 19:2025.02.19.639017. doi: 10.1101/2025.02.19.639017. bioRxiv. 2025. PMID: 40027745 Free PMC article. Preprint.
-
The Batalogue: an overview of betacoronaviruses with future pandemic potential.FEMS Microbiol Rev. 2025 Jan 14;49:fuaf023. doi: 10.1093/femsre/fuaf023. FEMS Microbiol Rev. 2025. PMID: 40434829 Free PMC article. Review.
-
Structures and receptor binding activities of merbecovirus spike proteins reveal key signatures for human DPP4 adaptation.Sci Adv. 2025 Jul 11;11(28):eadv7296. doi: 10.1126/sciadv.adv7296. Epub 2025 Jul 11. Sci Adv. 2025. PMID: 40644548 Free PMC article.
-
HKU5 bat merbecoviruses engage bat and mink ACE2 as entry receptors.Nat Commun. 2025 Jul 24;16(1):6822. doi: 10.1038/s41467-025-61583-7. Nat Commun. 2025. PMID: 40707428 Free PMC article.
-
ACE2 utilization of HKU25 clade MERS-related coronaviruses with broad geographic distribution.Res Sq [Preprint]. 2025 Mar 13:rs.3.rs-6097445. doi: 10.21203/rs.3.rs-6097445/v1. Res Sq. 2025. PMID: 40162213 Free PMC article. Preprint.
References
-
- Zaki, A. M., van Boheemen, S., Bestebroer, T. M., Osterhaus, A. D. & Fouchier, R. A. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N. Engl. J. Med.367, 1814–1820 (2012). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

