Clinico-genomic study reveals association of dengue virus genome high frequency mutations with dengue disease severity
- PMID: 40436903
- PMCID: PMC12120001
- DOI: 10.1038/s41598-025-00462-z
Clinico-genomic study reveals association of dengue virus genome high frequency mutations with dengue disease severity
Abstract
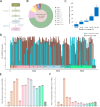
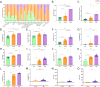
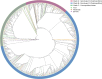
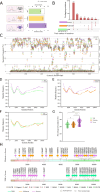
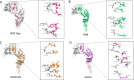
Transmission of the dengue virus (DENV) places a huge burden on public health in several endemic regions. Like other RNA viruses, mutations in the DENV genome greatly governs its virulence, transmissibility, and interaction with the host immune system. Present study focuses on integrated analysis of mutation and clinical data accompanied at the onset of dengue fever. The findings from the associated clinical data with the variants of the DENV are critical for early detection of the disease and understanding the disease progression. RNA was isolated from the 1310 serum samples collected from the NS1-antigen positive dengue patients. Serotyping reveals that DENV-2 was predominant in circulation. The genome of 1305 DENV-2 was sequenced using Oxford Nanopore Technology and Illumina platforms. A total of 1023 DENV-2 demonstrated > 50% genome coverage. Mutation analysis across the 1023 DENV-2 genomes yielded a total of 2667 mutations including 627 non-synonymous and 2040 synonymous mutations. We observed a notable over-representation of synonymous mutations in prM and ancC genes while a higher occurrence of non-synonymous mutations was found in ancC, prM, and M proteins. Comparison of mutation frequency between mild and severe demonstrates higher mutation frequency in severe phenotype. Moreover, we observed a total of 56 significant mutations including 23 in severe, 17 in moderate and 16 in mild. The E-protein having non-synonymous mutations were docked with DC-SIGN with lower binding energy (ΔG = - 11.9 kcal/mol) for severe as compared to mild (ΔG = - 13.5 kcal/mol), suggesting lesser affinity of E-protein and DC-SIGN in case of severe as compared to the mild. We have identified the core set of high frequency mutations significantly associated with distinct dengue disease severity viz., mild, moderate and severe. Furthermore, in-silico protein modelling and docking studies demonstrate the potential functional role of the non-synonymous mutations identified across E-protein in severe dengue.
Keywords: Dengue; Disease severity; Leukopenia; Mutation analysis; Thrombocytopenia.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Competing interests: The authors declare no competing interests. Ethics statement: The study was designed in accordance with the Declaration of Helsinki and was approved by the institutional ethics committee of Council of Scientific & Industrial Research-Institute of Genomics and Integrative Biology, Delhi, India (Ref No: CSIR-IGIB/IHEC/2020–21/01). The patients/participants provided their written informed consent before participation in this study.
Figures
Similar articles
-
Clinical characteristics and serotype association of dengue and dengue like illness in Pakistan.PLoS Negl Trop Dis. 2025 May 12;19(5):e0012978. doi: 10.1371/journal.pntd.0012978. eCollection 2025 May. PLoS Negl Trop Dis. 2025. PMID: 40354421 Free PMC article.
-
Haematological parameters as predictors of severe dengue: A study from northern districts of West Bengal, India.J Vector Borne Dis. 2025 Apr 1;62(2):218-225. doi: 10.4103/JVBD.JVBD_119_24. Epub 2024 Dec 5. J Vector Borne Dis. 2025. PMID: 39636263
-
Simultaneous Cocirculation of 2 Genotypes of Dengue Virus Serotype 3 Causing a Large Outbreak in Sri Lanka in 2023.J Infect Dis. 2025 Apr 15;231(4):1041-1048. doi: 10.1093/infdis/jiae474. J Infect Dis. 2025. PMID: 39387651
-
Meta-Analysis of Dengue Severity during Infection by Different Dengue Virus Serotypes in Primary and Secondary Infections.PLoS One. 2016 May 23;11(5):e0154760. doi: 10.1371/journal.pone.0154760. eCollection 2016. PLoS One. 2016. PMID: 27213782 Free PMC article.
-
Systemic pharmacological treatments for chronic plaque psoriasis: a network meta-analysis.Cochrane Database Syst Rev. 2021 Apr 19;4(4):CD011535. doi: 10.1002/14651858.CD011535.pub4. Cochrane Database Syst Rev. 2021. Update in: Cochrane Database Syst Rev. 2022 May 23;5:CD011535. doi: 10.1002/14651858.CD011535.pub5. PMID: 33871055 Free PMC article. Updated.
References
-
- Introduction to Dengue OpenWHO. https://openwho.org/courses/dengue-introduction.
-
- Dengue and severe dengue. https://www.who.int/en/news-room/fact-sheets/detail/dengue-and-severe-de....
-
- DENGUE SITUATION IN INDIA : National Center for Vector Borne Diseases Control (NCVBDC). https://ncvbdc.mohfw.gov.in/index4.php?lang=1&level=0&linkid=431&lid=3715.
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

