Sulfonohydrazide as a potential inhibitor of SARS-CoV-2 infection
- PMID: 40437072
- PMCID: PMC12119938
- DOI: 10.1038/s41598-025-03685-2
Sulfonohydrazide as a potential inhibitor of SARS-CoV-2 infection
Abstract
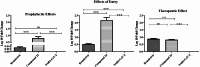
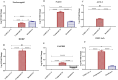
The COVID-19 pandemic caused immense mortality and morbidity reporting 704,753,890 cases worldwide. The repercussions of this pandemic are still being felt in the form of newly evolving variants and infections. The pandemic has pointed towards the need for the development of new and effective agents against SARS-CoV-2 infection. Sulfonohydrazides are a class of compounds with a wide range of therapeutic potential. The present study aims to identify the anti-SARS-CoV-2 potential of Sulfonohydrazide compounds. Twenty-five Sulfonohydrazides derivatives were evaluated for anti-viral potential via plaque reduction assay (PRA) and cytopathic effect (CPE) analysis in-vitro. Treatment point assay was employed for the strategic evaluation of antiviral compound at the particular stages of the SARS-CoV-2 life cycle. Gene expression analysis was also carried out, which was supported by immunofluorescence assays targeting the N and S proteins of SARS-CoV-2, alongside fold-change analysis, to identify a robust and multifaceted approach for the understanding of viral dynamics. Moreover, ligand-inhibitor interactions were assessed by in- silico studies. Compound 24 (4(E)-4-methyl-N'-(2,3,4-trihydroxybenzylidene)benzenesulfonohydrazide) was identified as the most potent molecule that inhibited SARS-CoV-2 infection (92.85 ± 3.57%) via PRA. The time point assay revealed that the effect of the compound might be at the entry point, which might be due to the down-regulation of the Spike (S) and Angiotensin-converting enzyme 2 (ACE-2) genes by the compound. The gene expression analysis of ORF1a/b by qRT-PCR indicated reduction in viral load after compound treatment, as indicated by a higher cycle threshold (Ct) value. Moreover, the compound 24 also downregulated the expression of S, RdRp, and ACE-2. Furthermore, the interaction of compound 24 with S, RdRp, and ACE-2 was predicted via molecular docking, which validated the interaction and possible anti-SARS-CoV-2 effect. Additionally, immunofluorescence staining analysis of spike and nucleocapsid proteins also showed downregulation in SARS-CoV-2 infected cells. Overall, the acquired data suggested that Sulfonohydrazide derivative 24 inhibits SARS-CoV-2 entry and replication.
Keywords: Angiotensin-converting enzyme 2; Antivirals; Multi-target therapeutics; RNA dependent RNA polymerase; Severe acute respiratory syndrome coronavirus 2; Spike; Sulfonohydrazides.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Competing interests: The authors declare no competing interests. Ethics approval and consent to participate: The research study was ethically approved by the Independent Ethics Committee (IEC), Dr. Panjwani Center for Molecular Medicine and & Drug Research, International Center for Chemical and Biological Sciences, University of Karachi (Reference # ICCBS/IEC-066-HNS/PC2021/Protocol/2.0). This study was conducted in accordance with the principles of the Declaration of Helsinki. All experiments were performed in accordance with relevant guidelines and regulations. Consent to participate: Informed consent for taking the nasal swab have been obtained from participant in accordance with relevant guidelines and regulations.
Figures




Similar articles
-
Multidisciplinary Approaches Identify Compounds that Bind to Human ACE2 or SARS-CoV-2 Spike Protein as Candidates to Block SARS-CoV-2-ACE2 Receptor Interactions.mBio. 2021 Mar 30;12(2):e03681-20. doi: 10.1128/mBio.03681-20. mBio. 2021. PMID: 33785634 Free PMC article.
-
Discovery and Evaluation of Entry Inhibitors for SARS-CoV-2 and Its Emerging Variants.J Virol. 2021 Nov 23;95(24):e0143721. doi: 10.1128/JVI.01437-21. Epub 2021 Sep 22. J Virol. 2021. PMID: 34550770 Free PMC article.
-
Identification of nitrile-containing isoquinoline-related natural product derivatives as coronavirus entry inhibitors in silico and in vitro.Biomed Pharmacother. 2024 Nov;180:117517. doi: 10.1016/j.biopha.2024.117517. Epub 2024 Oct 1. Biomed Pharmacother. 2024. PMID: 39357326
-
Inhibition of S-protein RBD and hACE2 Interaction for Control of SARSCoV- 2 Infection (COVID-19).Mini Rev Med Chem. 2021;21(6):689-703. doi: 10.2174/1389557520666201117111259. Mini Rev Med Chem. 2021. PMID: 33208074 Review.
-
Flavonols as potential antiviral drugs targeting SARS-CoV-2 proteases (3CLpro and PLpro), spike protein, RNA-dependent RNA polymerase (RdRp) and angiotensin-converting enzyme II receptor (ACE2).Eur J Pharmacol. 2021 Jan 15;891:173759. doi: 10.1016/j.ejphar.2020.173759. Epub 2020 Nov 27. Eur J Pharmacol. 2021. PMID: 33249077 Free PMC article. Review.
References
-
- Lamers, M. M. & Haagmans, B. L. SARS-CoV-2 pathogenesis. Nat. Rev. Microbiol.20 (5), 270–284 (2022). - PubMed
-
- WHO. Coronavirus disease (COVID-19) pandemic. World Health Organization, Geneva. https://www.who.int/emergencies/diseases/novel-coronavirus-2019 (2020).
-
- Cascella, M. et al. Features, evaluation, and treatment of coronavirus (COVID-19) [Updated 2023 Aug 18]. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; Jan (2024). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

