Two distinct host-specialized fungal species cause white-nose disease in bats
- PMID: 40437097
- PMCID: PMC12222008
- DOI: 10.1038/s41586-025-09060-5
Two distinct host-specialized fungal species cause white-nose disease in bats
Abstract
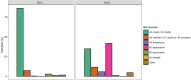
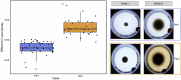
The emergence of infectious diseases, particularly those caused by fungal pathogens, poses serious threats to public health, wildlife and ecosystem stability1. Host-fungus interactions and environmental factors have been extensively examined2-4. However, the role of genetic variability in pathogens is often less well-studied, even for diseases such as white-nose in bats, which has caused one of the highest disease-driven death tolls documented in nonhuman mammals5. Previous research on white-nose disease has primarily focused on variations in disease outcomes attributed to host traits or environmental conditions6-8, but has neglected pathogen variability. Here we leverage an extensive reference collection of 5,479 fungal isolates from 27 countries to reveal that the widespread causative agent is not a single species but two sympatric cryptic species, each exhibiting host specialization. Our findings provide evidence of recombination in each species, but significant genetic differentiation across their genomes, including differences in genome organization. Both species contain geographically differentiated populations, which enabled us to identify the species introduced to North America and trace its source population to a region in Ukraine. In light of our discovery of the existence of two cryptic species of the causative agent of white-nose disease, our research underscores the need to integrate the study of pathogen variability into comprehensive disease surveillance, management and prevention strategies. This holistic approach is crucial for enhancing our understanding of diseases and implementing effective measures to prevent their spread.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures











References
-
- Knogge, W. Molecular basis of specificity in host/fungus interactions. Eur. J. Plant Pathol.102, 807–816 (1996).
-
- Rödder, D., Veith, M. & Lötters, S. Environmental gradients explaining the prevalence and intensity of infection with the amphibian chytrid fungus: the host’s perspective. Anim. Conserv.11, 513–517 (2008).
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

