Structure-informed design of an ultrabright RNA-activated fluorophore
- PMID: 40437193
- PMCID: PMC12329784
- DOI: 10.1038/s41557-025-01832-w
Structure-informed design of an ultrabright RNA-activated fluorophore
Abstract
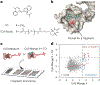
RNA-based fluorogenic aptamers, such as Mango, are uniquely powerful tools for imaging RNA that activate the fluorescence of a weakly or non-fluorescent small molecule when bound. A central challenge has been to develop brighter, more specific and high-affinity aptamer-ligand systems for cellular imaging. Here we report an ultrabright fluorophore for the Mango II system discovered using a structure-informed, fragment-based small-molecule microarray approach. This dye-termed SALAD1 (structure-informed, array-enabled LigAnD 1)-exhibits subnanomolar aptamer affinity and 3.5-fold brighter fluorescence than Mango II-TO1-biotin pair, a widely used fluorogenic system. Performance was improved by modulating RNA-dye molecular recognition without altering the fluorophore's π-system. High-resolution X-ray structures reveal the binding mode for SALAD1, which exhibits improved pocket occupancy, a more defined binding pose and a unique bonding interaction with potassium. SALAD1 is cell-permeable and facilitates improved in-cell confocal RNA imaging. This work introduces an additional RNA-activated fluorophore demonstrating how fragment-based ligand discovery can be used to create high-performance ligands for RNA targets.
© 2025. This is a U.S. Government work and not under copyright protection in the US; foreign copyright protection may apply.
Conflict of interest statement
Competing interests: K.M.W. is an advisor to and holds equity in Ribometrix, ForagR Medicines and A-Form Solutions. The remaining authors declare no competing interests.
Figures
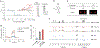
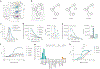
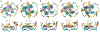

Update of
-
Structure-Informed Design of an Ultra Bright RNA-activated Fluorophore.Res Sq [Preprint]. 2024 Aug 5:rs.3.rs-4750449. doi: 10.21203/rs.3.rs-4750449/v1. Res Sq. 2024. Update in: Nat Chem. 2025 Aug;17(8):1188-1195. doi: 10.1038/s41557-025-01832-w. PMID: 39149476 Free PMC article. Updated. Preprint.
Similar articles
-
Structure-Informed Design of an Ultra Bright RNA-activated Fluorophore.Res Sq [Preprint]. 2024 Aug 5:rs.3.rs-4750449. doi: 10.21203/rs.3.rs-4750449/v1. Res Sq. 2024. Update in: Nat Chem. 2025 Aug;17(8):1188-1195. doi: 10.1038/s41557-025-01832-w. PMID: 39149476 Free PMC article. Updated. Preprint.
-
Symmetry breaking of fluorophore binding to a G-quadruplex generates an RNA aptamer with picomolar KD.Nucleic Acids Res. 2024 Aug 12;52(14):8039-8051. doi: 10.1093/nar/gkae493. Nucleic Acids Res. 2024. PMID: 38945550 Free PMC article.
-
Structural basis for ring-opening fluorescence by the RhoBAST RNA aptamer.Nucleic Acids Res. 2025 Jun 20;53(12):gkaf555. doi: 10.1093/nar/gkaf555. Nucleic Acids Res. 2025. PMID: 40598898 Free PMC article.
-
Signs and symptoms to determine if a patient presenting in primary care or hospital outpatient settings has COVID-19.Cochrane Database Syst Rev. 2022 May 20;5(5):CD013665. doi: 10.1002/14651858.CD013665.pub3. Cochrane Database Syst Rev. 2022. PMID: 35593186 Free PMC article.
-
Management of urinary stones by experts in stone disease (ESD 2025).Arch Ital Urol Androl. 2025 Jun 30;97(2):14085. doi: 10.4081/aiua.2025.14085. Epub 2025 Jun 30. Arch Ital Urol Androl. 2025. PMID: 40583613 Review.
References
-
- Yin P, Kuang S & Nie Z Fluorescent RNA tags for in situ RNA imaging in living cells. Anal. Sens 3, e202200090 (2023).
-
- Armitage BA Imaging of RNA in live cells. Curr. Opin. Chem. Biol 15, 806–812 (2011). - PubMed
MeSH terms
Substances
Grants and funding
- ZIAHL006188/U.S. Department of Health & Human Services | NIH | National Heart, Lung, and Blood Institute (NHLBI)
- ZIA HL006188/ImNIH/Intramural NIH HHS/United States
- R35 GM122532/GM/NIGMS NIH HHS/United States
- ZIA BC011585/ImNIH/Intramural NIH HHS/United States
- 1ZIABC011585-08/U.S. Department of Health & Human Services | NIH | National Cancer Institute (NCI)
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

