InterpolAI: deep learning-based optical flow interpolation and restoration of biomedical images for improved 3D tissue mapping
- PMID: 40437217
- PMCID: PMC12240858
- DOI: 10.1038/s41592-025-02712-4
InterpolAI: deep learning-based optical flow interpolation and restoration of biomedical images for improved 3D tissue mapping
Abstract
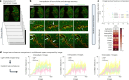
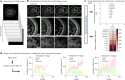
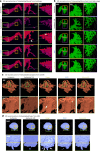
Recent advances in imaging and computation have enabled analysis of large three-dimensional (3D) biological datasets, revealing spatial composition, morphology, cellular interactions and rare events. However, the accuracy of these analyses is limited by image quality, which can be compromised by missing data, tissue damage or low resolution due to mechanical, temporal or financial constraints. Here, we introduce InterpolAI, a method for interpolation of synthetic images between pairs of authentic images in a stack of images, by leveraging frame interpolation for large image motion, an optical flow-based artificial intelligence (AI) model. InterpolAI outperforms both linear interpolation and state-of-the-art optical flow-based method XVFI, preserving microanatomical features and cell counts, and image contrast, variance and luminance. InterpolAI repairs tissue damages and reduces stitching artifacts. We validated InterpolAI across multiple imaging modalities, species, staining techniques and pixel resolutions. This work demonstrates the potential of AI in improving the resolution, throughput and quality of image datasets to enable improved 3D imaging.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures





Update of
-
Generative interpolation and restoration of images using deep learning for improved 3D tissue mapping.bioRxiv [Preprint]. 2024 Mar 28:2024.03.07.583909. doi: 10.1101/2024.03.07.583909. bioRxiv. 2024. Update in: Nat Methods. 2025 Jul;22(7):1556-1567. doi: 10.1038/s41592-025-02712-4. PMID: 38496512 Free PMC article. Updated. Preprint.
References
-
- Forjaz, A. et al. Three-dimensional assessments are necessary to determine the true spatial tissue composition of diseased tissues. Preprint bioRxiv10.1101/2023.12.04.569986 (2023).
-
- Cline, H. E., Dumoulin, C. L., Hart, H. R., Lorensen, W. E. & Ludke, S. 3D reconstruction of the brain from magnetic resonance images using a connectivity algorithm. Magn. Reson. Imaging5, 345–352 (1987). - PubMed
MeSH terms
Grants and funding
- U54 AR081774/AR/NIAMS NIH HHS/United States
- R24 MH114785/MH/NIMH NIH HHS/United States
- UH3 CA275681/CA/NCI NIH HHS/United States
- UG3CA275681/U.S. Department of Health & Human Services | NIH | National Institute of Allergy and Infectious Diseases (NIAID)
- R24MH114785/U.S. Department of Health & Human Services | NIH | National Institute of Mental Health (NIMH)
- U54AR081774/U.S. Department of Health & Human Services | NIH | National Institute of Allergy and Infectious Diseases (NIAID)
- U54 CA268083/CA/NCI NIH HHS/United States
- U54CA268083/U.S. Department of Health & Human Services | NIH | National Cancer Institute (NCI)
- UG3 CA275681/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources

