Molecular determinants of sotorasib clinical efficacy in KRASG12C-mutated non-small-cell lung cancer
- PMID: 40437272
- PMCID: PMC12353874
- DOI: 10.1038/s41591-025-03732-5
Molecular determinants of sotorasib clinical efficacy in KRASG12C-mutated non-small-cell lung cancer
Abstract
Molecular determinants of KRAS(G12C)inhibitor efficacy in KRASG12C-mutated non-small-cell lung cancer (NSCLC) remain poorly characterized. Here we report one of the largest integrated analyses to date of sotorasib clinical efficacy biomarkers from the phase 2 CodeBreaK 100 and phase 3 CodeBreaK 200 studies. We reveal differential sotorasib activity and relative benefit compared to docetaxel across KRASG12C-mutated NSCLC co-mutational subsets and transcriptional subtypes. We also identify low expression of TTF1 and KEAP1 co-mutations/NRF2 activation as major determinants of sotorasib anti-tumor efficacy and adverse prognostic features. Exploratory analyses highlight potential tumor cell-extrinsic contributors to sotorasib anti-tumor activity and suggest that early on-treatment clearance of KRASG12C- circulating tumor DNA may refine clinical response prediction algorithms. Our findings advance precision medicine for patients with KRASG12C-mutated NSCLC and establish a framework for patient stratification and selection for treatment intensification with rationally applied therapeutic combinations.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: F.S. reports consulting/advisory board fees from Amgen, AstraZeneca, AImmune (spouse), BeiGene, BergenBio, BridgeBio, Bristol Myers Squibb, Calithera Biosciences, Guardant Health, Hookipa Pharma, Merck Sharp & Dohme, Novartis, Novocure, Regeneron, Revolution Medicines, Roche and Tango Therapeutics; grant or research support (to institution) from Amgen, Mirati Therapeutics, Revolution Medicines, Pfizer, Novartis and Merck & Co; stock ownership in BioNTech (ended in 2021) and Moderna (ended in 2021); honoraria/lecture fees from the European Society for Medical Oncology (ESMO), the Japanese Lung Cancer Society, Medscape, Intellisphere, VSPO McGill University, RV Mais Promocao Eventos, MJH Life Sciences, IDEOlogy Health, MI&T, Physiciansʼ Education Resource (PER), Curio, DAVA Oncology, the American Association for Cancer Research (AACR) and the International Association for the Study of Lung Cancer (IASLC); and travel support from DAVA Oncology, Tango Therapeutics, AstraZeneca, Amgen, Inc., Bristol Myers Squibb, Revolution Medicines, AACR, IASLC, MJH Life Sciences, IDEOlogy Health, MI&T, PER and Curio. B.T.L. has received research funding from Roche/Genentech (to institution), AstraZeneca (to institution), Daiichi Sankyo (to institution), Hengrui Therapeutics (to institution), Amgen (to institution), Eli Lilly (to institution), MORE Health (to institution), Bolt Biotherapeutics (to institution) and Ambrx (to institution); has patents, royalties and other intellectual property from US62/514,661 (to institution), US62/685,057 (to institution), Karger Publishers and Shanghai Jiao Tong University Press; has received travel, accommodations and expenses from MORE Health and Jiangsu Hengrui Medicine; and has uncompensated relationships with Amgen, AstraZeneca, Genentech, Eli Lilly, Boehringer Ingelheim and Daiichi Sankyo. A.J.d.L. has financial interests in and has received institutional and research grant funding from Bristol Myers Squibb, Merck Sharp & Dohme, Boehringer Ingelheim and AstraZeneca and has non-financial and other interests in Merck Serono and Roche. D.S.H. owns stock or has other ownership interests in OncoResponse, Telperian and MolecularMatch; has acted in a consulting or advisory role for Bayer, Guidepoint Global, Gerson Lehrman Group, Alphasights, Axiom Biotechnologies, Medscape, Numab, Pfizer, Takeda, Trieza Therapeutics, WebMD, Infinity Pharmaceuticals, Amgen, Adaptimmune, Boxer Capital, EcoR1 Capital, Tavistock Life Sciences, Baxter, COG, Genentech, GroupH, Janssen, Acuta, HCW Precision, Prime Oncology, ST Cube, Alkermes, AUM Biosciences, BridgeBio, Cor2Ed, Gilead Sciences, Immunogen, Liberum, Oncologia Brasil, Pharma Intelligence, Precision Oncology Experimental Therapeutics, Turning Point Therapeutics, ZIOPHARM Oncology, Cowen, Gennao Bio, MedaCorp, YingLing Pharma and RAIN; received research funding from Genentech (to institution), Amgen (to institution), Daiichi Sankyo (to institution), Adaptimmune (to institution), AbbVie (to institution), Bayer (to institution), Infinity Pharmaceuticals (to institution), Kite, a Gilead Company (to institution), MedImmune (to institution), the National Cancer Institute (to institution), Fate Therapeutics (to institution), Pfizer (to institution), Novartis (to institution), Numab (to institution), Turning Point Therapeutics (to institution), Kyowa (to institution), Loxo (to institution), Merck (to institution), Eisai (to institution), Genmab (to institution), Mirati Therapeutics (to institution), Mologen (to institution), Takeda (to institution), AstraZeneca (to institution), Navire (to institution), VM Pharma (to institution), Erasca, Inc. (to institution), Bristol Myers Squibb (to institution), Adlai Nortye (to institution), Seagen (to institution), Deciphera (to institution), Pyramid Biosciences (to institution), Eli Lilly (to institution), Endeavor BioMedicines (to institution), F. Hoffmann LaRoche (to institution), Ignyta (to institution), Teckro (to institution) and TCR2 Therapeutics (to institution); and received reimbursement for travel, accommodations, expenses from Genmab, the Society for Immunotherapy of Cancer, Bayer Schering Pharma, the American Society of Clinical Oncology, AACR and Telperian. H.L. has financial interests in or received personal and other fees from Daiichi Sankyo, AstraZeneca, Pfizer, Novartis, Amgen, Merck Sharp & Dohme, Roche, Bristol Myers Squibb, Eli Lilly and Boehringer Ingelheim. J.W. has financial interests in and/or received personal, advisory board or lectures fees or institutional/research grants from Amgen, AstraZeneca, Bayer, Blueprint, Bristol Myers Squibb, Boehringer Ingelheim, Chugai, Daiichi Sankyo, Merck, Janssen, Eli Lilly, Loxo, Merck Sharp & Dohme, Novartis, Pfizer, Roche, Seattle Genetics, Takeda, Turning Point and Nuvalent. G.K.D. has acted in a consulting or advisory role for AstraZeneca, Mirati Therapeutics, Eli Lilly and Amgen and received research funding from Amgen (to institution), AstraZeneca (to institution), Mirati Therapeutics (to institution), Eli Lilly (to institution), Sanofi (to institution), Bioatla (to institution), Regeneron (to institution), Iovance Biotherapeutics (to institution) and Revolution Medicines (to institution). A.C.F. has acted in a consulting or advisory role for Amgen, AstraZeneca, Boehringer Ingelheim, Bristol Myers Squibb, Daichii Sankyo, Janssen, Medscape, Merck Sharp & Dohme, Roche/Genentech and Takeda; presented in a company-organized public event for Amgen, AstraZeneca, Bristol Myers Squibb, Foundation Medicine, Medscape, Merck Sharp & Dohme, Roche/Genentech and Takeda; and received grants or research support as a sub-investigator in trials (institutional financial support for clinical trials) sponsored by Amgen, AstraZeneca, Boehringer Ingelheim, Bristol Myers Squibb, Merck Sharp & Dohme and Roche/Genentech. P.T. has acted in a consulting or advisory role for AstraZeneca, Roche, Bristol Myers Squibb Foundation, Takeda, Amgen and Janssen and has received reimbursement for travel, accommodations and expenses from Bristol Myers Squibb/Pfizer, AstraZeneca and Takeda. V.V. has received honoraria from ITeos Therapeutics; acted in a consulting or advisory role for Bristol Myers Squibb, Merck, AstraZeneca/MedImmune, GlaxoSmithKline, Amgen, Elevation Oncology, Merus and Taiho Oncology; and received research funding from Genentech (to institution), Trovagene (to institution), Eisai (to institution), OncoPlex Diagnostics (to institution), Alkermes (to institution), NantWorks (to institution), Genoptix (to institution), Altor BioScience (to institution), Merck (to institution), Bristol Myers Squibb (to institution), Atreca (to institution), Heat Biologics (to institution), Leap Therapeutics (to institution), RSIP Vision (to institution) and GlaxoSmithKline (to institution). A.J.v.d.W.: financial interests—institutional: research grants from AstraZeneca, Boehringer Ingelheim, Pfizer, Roche and Takeda; financial interests—fees to institution from AstraZeneca, Boehringer Ingelheim, Pfizer, Roche, Takeda, Janssen Cilag, Eli Lilly, Amgen and Merck. C.D. has no competing interests to disclose. L.P.-A.R. has financial interests in and/or has received research grants from and/or payment/honoraria for lectures, presentations and speakers bureau participation from Merck Sharp & Dohme, AstraZeneca, Pfizer, Bristol Myers Squibb, Eli Lilly, Roche, PharmaMar, Merck, Novartis, Servier, Amgen, Sanofi, Bayer, Mirati Therapeutics, GlaxoSmithKline, Janssen, Takeda and Daiichi Sankyo. G.M. has financial interests in and/or received personal, consulting, honoraria, lecture/presentation, speakers bureaus, writing, traveling or other fees from Roche Hellas, Novartis Greece, BMS Greece, MSD Greece, AstraZeneca Greece, Takeda Hellas, Janssen Greece, GSK Greece, Amgen Hellas, Sanofi Greece, Boehringer Greece and Pierre Fabre Greece; had a leadership or fiduciary role, unpaid, for ESMO working groups (Educational Publication Working Group and Adolescents and Young Adults Working Group); and has acted as institutional/principal investigator for Roche Hellas, Novartis Greece, BMS Greece, MSD Greece, AstraZeneca Greece, Gilead Greece, GSK Greece, Amgen Hellas and Sanofi Greece. A.S. has received research funding from AstraZeneca, Genentech/Roche and Bristol Myers Squibb and has uncompensated relationships with Genentech/Roche and AstraZeneca. E.N. has received honoraria from Apollomics, Roche/Genentech and Transgene; acted in a consulting or advisory role for Amgen, AstraZeneca, Bayer, BeiGene, Boehringer Ingelheim, Bristol Myers Squibb, Daiichi Sankyo Europe GmbH, Genmab, Janssen Oncology, Eli Lilly, Merck Serono, Merck Sharp & Dohme, Pfizer, Pierre Fabre, Qiagen, Roche, Sanofi and Takeda; received research funding (to institution) from Bristol Myers Squibb, Merck Serono, Pfizer and Roche; and received travel, accommodations and expenses from Bristol Myers Squibb, Eli Lilly, Merck Sharp & Dohme, Pfizer, Roche and Takeda. S.C. has received grants and contracts from Adene, AstraZeneca, BD Biosciences, Bristol Myers Squibb, Canon, Chugai, I-Medica, Janssen, Eli Lilly, Merck Sharp & Dohme, Pfizer, PharmaMar, Pierre Fabre, Regeneron, Roche, Sanofi, Takeda, Transdiag and Volition; received payment for expert testimony from Amgen, AstraZeneca, Bristol Myers Squibb, Boehringer Ingelheim, Chugai, Fabentech, Health Event, Janssen, MaaT Pharma, Merck Sharp & Dohme, Novartis, Pierre Fabre, Roche, Sanofi and Takeda; received support for attending meetings and/or travel from Amgen, AstraZeneca, Bristol Myers Squibb, Chiesi, Janssen, Laidet, Pfizer, Roche and Takeda; had a leadership or fiduciary role in other board, society, committee or advocacy groups, unpaid, from Adene, Association de Recherche et d’Information Scientifique en Oncologie Thoracique (ARISTOT), Ensemble Nous Poumons, the Lung Cancer Policy Network and the Société de Pneumologie de Langue Française; and has other financial or non-financial interests from the Société Nationale des Chemins de fer Français (SNCF) (consulting physician, paid). S.-W.K. has no competing interests to disclose. K.O. owns stock or has other ownership interests in Carpe Vitae Pharmaceuticals, DGC Diagnostics and RepLuca Pharmaceuticals; has received honoraria from Amgen, AstraZeneca, BeiGene, Boehringer Ingelheim, Bristol Myers Squibb, Ipsen, Diacchi, Merck Group, Merck Sharp & Dohme, Pfizer/EMD Serono, Roche, Seagen, Takeda and TriStar Technology Group; has acted in a consulting or advisory role for Amgen, AstraZeneca/MedImmune, BeiGene, Boehringer Ingelheim, Bristol Myers Squibb, Diacchi, Ipsen, Merck Sharp & Dohme, Pfizer, Roche/Genentech, Sanofi and Seagen; has participated in speakers bureaus for BeiGene, Boehringer Ingelheim, Bristol Myers Squibb, Janssen-Cilag, Merck Group, Merck Sharp & Dohme, Pfizer, Roche and Seagen; has received travel, accommodations and expenses from Bayer Holding and Sanofi; and is named on four active patents (institution)—two published and two provisional. D.R. has no competing interests to disclose. R.T. has received honoraria from AstraZeneca, Bristol Myers Squibb Japan, Chugai Pharma, Eli Lilly Japan, Merck Sharp & Dohme, Nippon Kayaku, Ono Pharmaceutical, Pfizer, Taiho Pharmaceutical, Takeda and Thermo Fisher Scientific and has received research funding from AbbVie (to institution), Amgen (to institution), AnHeart Therapeutics (to institution), Daiichi Sankyo (to institution), Eli Lilly Japan (to institution), Novartis (to institution), Pfizer (to institution) and Takeda (to institution). I.C. has no competing interests to disclose. C.R.L. has financial interests in and has received personal, institutional, advisory board, research grant and other fees from Amgen, Qiagen and Revolution Medicines and has non-financial relationships with Amgen, BI, Mirati Therapeutics, Revolution Medicines, Roche and Apollomics. A.H. is an employee and stockholder/shareholder in Amgen. L.M. is an employee and stockholder/shareholder in Amgen. T.W. is an employee and stockholder/shareholder in Amgen. A.A. is an employee and stockholder/shareholder in Amgen and is listed as an inventor on several Amgen patents (does not receive royalties on these patents). C.A.-A. is an employee and stockholder/shareholder in Amgen. A.P. is an employee and stockholder/shareholder in Amgen. A.R. is an employee and stockholder/shareholder in Amgen. B.M. is an employee and stockholder/shareholder in Amgen. M.S. has received institutional research grants from AstraZeneca, Bristol Myers Squibb and Johnson & Johnson as well as consulting fees and fees of CME presentations and has participated on advisory boards for Amgen, AstraZeneca, Bristol Myers Squibb, Gilead, GlaxoSmithKline, Immunocore, Johnson & Johnson, Merck Sharp & Dohme, Novartis, Regeneron, Roche and Sanofi.
Figures
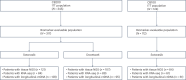
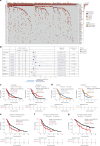

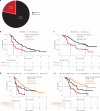

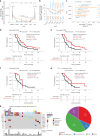

References
-
- Finn, S. P. et al. Prognostic impact of KRAS G12C mutation in patients with NSCLC: results from the European Thoracic Oncology Platform Lungscape Project. J. Thorac. Oncol.16, 990–1002 (2021). - PubMed
-
- Canon, J. et al. The clinical KRAS(G12C) inhibitor AMG 510 drives anti-tumour immunity. Nature575, 217–223 (2019). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

