Genetic engineering of E. coli K-12 for heterologous carbohydrate antigen production
- PMID: 40437542
- PMCID: PMC12121013
- DOI: 10.1186/s12934-025-02749-2
Genetic engineering of E. coli K-12 for heterologous carbohydrate antigen production
Abstract
Background: Carbohydrate-based vaccines have made a remarkable impact on public health over the past three decades. Efficient production of carbohydrate antigens is a crucial prerequisite for the development of such vaccines. The enzymes involved in the synthesis of bacterial surface carbohydrate antigens are usually encoded by large, uninterrupted gene clusters. Non-pathogenic E. coli glycoengineering starts with the genetic manipulation of these clusters. Heterologous gene cluster recombination through an expression plasmid has several drawbacks, including continuous antibiotic selection pressure, genetic instability, and metabolic burdens. In contrast, chromosome-level gene cluster expression can minimize the metabolic effects on the host and reduce industrial costs.
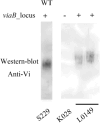
Results: In this study, we employed the suicide vector-mediated allelic exchange method to directly replace the native polysaccharide gene clusters in E. coli with heterologous ones. Unlike previously strategies, this method does not rely on I-SceI endonuclease or CRISPR/Cas system to release the linearized DNA insert and λ-red recombinase to promote its homologous recombination. Meanwhile, the vectors could be conveniently constructed by assembling multiple large DNA fragments in order in vitro. The scarless chromosomal insertions were confirmed by whole-genome sequencing and the polysaccharide phenotypes of all glycoengineered E. coli mutants were evaluated through growth curves, silver staining, western blot, and flow cytometry. The data indicated that there was no obvious metabolic burden associated with the insertion of large gene clusters into the E. coli W3110 O-antigen locus, and the glycoengineered E. coli can produce LPS with a recovery rate around 1% of the bacterial dry weight. Moreover, the immunogenicity of the heterologously expressed carbohydrate antigens was analyzed by mice immunization experiments. The ELISA data demonstrated the successful induction of anti-polysaccharide IgM or IgG antibodies.
Conclusions: We have provided a convenient and reliable genomic glycoengineering method to produce efficacious, durable, and cost-effective carbohydrate antigens in non-pathogenic E. coli. Non-pathogenic E. coli glycoengineering has great potential for the highly efficient synthesis of heterologous polysaccharides and can serve as a versatile platform to produce next-generation biomedical agents, including glycoconjugate vaccines, glycoengineered minicells or outer membrane vesicles (OMVs), polysaccharide-based diagnostic reagents, and more.
Keywords: E. coli K-12; S. enterica; DNA Long fragment editing; Glycoengineering; Polysaccharide biosynthesis.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: Animal experiments were conducted in compliance with the Animal Welfare Act and regulations stated in the Guide for the Care and Use of Laboratory Animals, which was approved by the Institutional Animal Care and Use Committee of Southwest University (Approval No. LAC2024-1-0187). Competing interests: The authors declare no competing interests.
Figures





References
-
- Whitfield C, Wear SS, Sande C. Assembly of bacterial capsular polysaccharides and exopolysaccharides. Annu Rev Microbiol. 2020;74:521–43. - PubMed
-
- Domínguez-Medina CC, Pérez-Toledo M, Schager AE, Marshall JL, Cook CN, Bobat S, Hwang H, Chun BJ, Logan E, Bryant JA, Channell WM, Morris FC, Jossi SE, Alshayea A, Rossiter AE, Barrow PA, Horsnell WG, MacLennan CA, Henderson IR, Lakey JH, Gumbart JC, López-Macías C, Bavro VN, Cunningham AF. Outer membrane protein size and LPS O-antigen define protective antibody targeting to the Salmonella surface. Nat Commun. 2020;11:851. - PMC - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

