Unexpected effect of an axial ligand mutation in the type 1 copper center in small laccase: structure-based analyses and engineering to increase reduction potential and activity
- PMID: 40438172
- PMCID: PMC12109610
- DOI: 10.1039/d5sc02177d
Unexpected effect of an axial ligand mutation in the type 1 copper center in small laccase: structure-based analyses and engineering to increase reduction potential and activity
Abstract
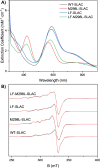
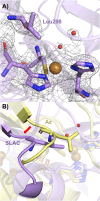
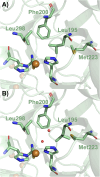
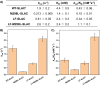
Type 1 copper (T1Cu) centers are crucial in biological electron transfer (ET) processes, exhibiting a wide range of reduction potentials to match their redox partners and optimize ET rates. While tuning in mononuclear T1Cu proteins like azurin has been successful, it is more difficult for multicopper oxidases. Specifically, while replacing axial methionine to leucine in azurin increased its by ∼100 mV, the corresponding M298L mutation in small laccase from Streptomyces coelicolor (SLAC) unexpectedly decreased its by 12 mV. X-ray crystallography revealed two axial water molecules in M298L-SLAC, leading to the decrease of due to decreased hydrophobicity. Structural alignment and molecular dynamics simulation indicated a key difference in T1Cu axial loop position, leading to the different outcome upon methionine to leucine mutation. Based on structural analyses, we introduced additional F195L and I200F mutations, leading to partial removal of axial waters, a 122-mV increase in , and a 7-fold increase in k cat/K M from M298L-SLAC. These findings highlight the complexity of tuning in multicopper oxidases and provide valuable insights into how structure-based protein engineering can contribute to the broader understanding of T1Cu center, and reactivity tuning for applications, such as in solar energy transfer, fuel cells, and bioremediation.
This journal is © The Royal Society of Chemistry.
Conflict of interest statement
There are no conflicts to declare.
Figures


Similar articles
-
Beyond Blue: Systematic Modulation of Electronic Structure and Redox Properties of Type 1 Copper in Azurin.J Am Chem Soc. 2025 Jul 16;147(28):24825-24837. doi: 10.1021/jacs.5c07009. Epub 2025 Jul 8. J Am Chem Soc. 2025. PMID: 40626760
-
Assessing the comparative effects of interventions in COPD: a tutorial on network meta-analysis for clinicians.Respir Res. 2024 Dec 21;25(1):438. doi: 10.1186/s12931-024-03056-x. Respir Res. 2024. PMID: 39709425 Free PMC article. Review.
-
Signs and symptoms to determine if a patient presenting in primary care or hospital outpatient settings has COVID-19.Cochrane Database Syst Rev. 2022 May 20;5(5):CD013665. doi: 10.1002/14651858.CD013665.pub3. Cochrane Database Syst Rev. 2022. PMID: 35593186 Free PMC article.
-
Exercise interventions and patient beliefs for people with hip, knee or hip and knee osteoarthritis: a mixed methods review.Cochrane Database Syst Rev. 2018 Apr 17;4(4):CD010842. doi: 10.1002/14651858.CD010842.pub2. Cochrane Database Syst Rev. 2018. PMID: 29664187 Free PMC article.
-
Maternal and neonatal outcomes of elective induction of labor.Evid Rep Technol Assess (Full Rep). 2009 Mar;(176):1-257. Evid Rep Technol Assess (Full Rep). 2009. PMID: 19408970 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Miscellaneous

