Immunological features of various molecular subtypes of cervical cancer and their prognostic implications in the context of disulfidptosis
- PMID: 40438679
- PMCID: PMC12116334
- DOI: 10.3389/fonc.2025.1574911
Immunological features of various molecular subtypes of cervical cancer and their prognostic implications in the context of disulfidptosis
Abstract
Objective: Cervical cancer ranks among the most prevalent malignancies impacting women globally. Disulfidptosis represents a recently identified pathway of cellular demise, although its role in the context of cervical cancer is not well elucidated. This research investigates the significance of Disulfidptosis-Related Genes (DRGs) within cervical cancer. Furthermore, it aims to analyze the differences in prognosis and immune infiltration among different molecular subtypes.
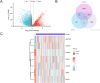
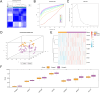
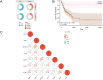
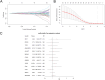
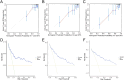
Methods: We compiled genes associated with cervical cancer and disulfidptosis from a variety of databases to perform a differential expression analysis. Subsequently, the samples are grouped through consensus clustering. To evaluate immune cell infiltration, we employed CIBERSORT. Additionally, immune checkpoint genes (ICGs) were gathered from existing literature and databases, enabling statistical analyses of two subtype samples of cervical cancer (CESC). Following our analyses using GO, KEGG, and GSEA to compare the differences between the two subtypes. Lastly, a prognostic risk model was constructed using LASSO regression and validated using ROC.
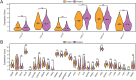
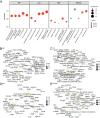
Results: This study identified seven key genes: PCBP3, ARNT, ANP32E, DSTN, CD2AP, EPAS1, and ACTN1.The consensus clustering analysis showed differences in immune cell infiltration and DFS(disease-free survival) among the various clusters. The immune checkpoint gene CXCL1 displayed highly significant statistical differences between subtype A (Cluster 1) and subtype B (Cluster 2) in cervical cancer (CESC) samples. The gene set enrichment analysis identified the negative regulation of peptidase activity and the IL-17 signaling pathway, which link to subtype-specific differentially expressed genes (DEGs).
Conclusion: Statistical analysis of the various subtypes of CESC samples highlighted the importance of subtype-specific therapeutic targets. Additionally, it seeks to enhance the accuracy of prognostic predictions, thereby establishing a foundation for the formulation of personalized treatment approaches.
Keywords: cervical cancer; disulfidptosis; immune cell infiltration; prognostic model; risk signature.
Copyright © 2025 Yao, Yang, Fu and Zhang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




References
LinkOut - more resources
Full Text Sources
Miscellaneous

