PRDM16, a new kid on the block in cardiovascular health and disease
- PMID: 40439125
- PMCID: PMC12310283
- DOI: 10.1093/cvr/cvaf089
PRDM16, a new kid on the block in cardiovascular health and disease
Abstract
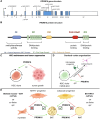
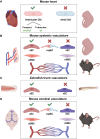
Transcriptional regulation is essential for the development, homeostasis, and function of all organisms. Transcription factors and epigenetic modifiers play an indispensable role by direct or indirect interaction with DNA or chromatin. Although the role of transcription factor PRDM16 in adipose, haematopoietic, skeletal, and neural cell lineage specification is well-documented, its function within the cardiovascular system has only recently gained significant attention. Similar as in adipose tissue, PRDM16 displays an asymmetric expression pattern within the cardiovascular system, where it is exclusively expressed by ventricular cardiomyocytes and endothelial and smooth muscle cells of arteries while being absent in their atrial and venous counterparts. Concordantly, an increasing number of clinical and preclinical studies have identified PRDM16 as an important multi-modal regulator of cardiovascular development and function. Moreover, aberrant PRDM16 expression has now been linked to (cardio)vascular diseases, including left ventricular non-compaction, migraine, and coronary artery disease. In this review, we give a synopsis of PRDM16's expression and function within (developing) cardiovascular tissues and provide insights into how impaired PRDM16 signalling contributes to cardiovascular disease.
Keywords: Cardiomyopathy; Cell-fate decision; PRDM16; Transcription factors; Vascular disease.
© The Author(s) 2025. Published by Oxford University Press on behalf of the European Society of Cardiology.
Conflict of interest statement
Conflict of interest: None declared.
Figures



References
-
- Nishikata I, Sasaki H, Iga M, Tateno Y, Imayoshi S, Asou N, Nakamura T, Morishita K. A novel EVI1 gene family, MEL1, lacking a PR domain (MEL1S) is expressed mainly in t(1;3)(p36;q21)-positive AML and blocks G-CSF-induced myeloid differentiation. Blood 2003;102:3323–3332. - PubMed
-
- Mochizuki N, Shimizu S, Nagasawa T, Tanaka H, Taniwaki M, Yokota J, Morishita K. A novel gene, MEL1, mapped to 1p36.3 is highly homologous to the MDS1/EVI1 gene and is transcriptionally activated in t(1;3)(p36;q21)-positive leukemia cells. Blood 2000;96:3209–3214. - PubMed
-
- Nishikata I, Nakahata S, Saito Y, Kaneda K, Ichihara E, Yamakawa N, Morishita K. Sumoylation of MEL1S at lysine 568 and its interaction with CtBP facilitates its repressor activity and the blockade of G-CSF-induced myeloid differentiation. Oncogene 2011;30:4194–4207. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- G099521N/2 Fonds voor Wetenschappelijk Onderzoek
- G060923N/2 Fonds voor Wetenschappelijk Onderzoek
- Fonds Spécial de Recherce
- 1C21200082/UCLouvain
- 1D21200068/UCLouvain
- 2 Belgian Heart Fund
- Belgian Society for Cardiology
- 2022-J1190990-225777/King Baudouin Foundation
- 2025-J1190990-0024422/King Baudouin Foundation
- BE 8635/1-1/German Research Foundation
- 1114125N/FWO
- 1S25817N/FWO
- 11P9W24N/FWO
- 1271824N/FWO
- Frans Van de Werf Fellowship for Clinical Cardiovascular Research 2024
LinkOut - more resources
Full Text Sources

