Polyphenol rewiring of the microbiome reduces methane emissions
- PMID: 40439232
- PMCID: PMC12203004
- DOI: 10.1093/ismejo/wraf108
Polyphenol rewiring of the microbiome reduces methane emissions
Abstract
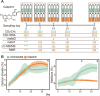
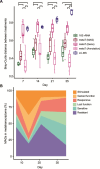
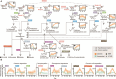
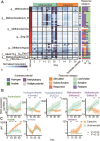
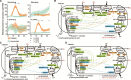
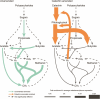
Methane mitigation is regarded as a critical strategy to combat the scale of global warming. Currently, ~40% of methane emissions originate from microbial sources, which is causing strategies to suppress methanogens-either through direct toxic effects or by diverting their substrates and energy-to gain traction. Problematically, current microbial methane mitigation knowledge lacks detailed microbiome-centered insights, limiting translation across conditions and ecosystems. Here we utilize genome-resolved metatranscriptomes and metabolomes to assess the impact of a proposed methane inhibitor, catechin, on greenhouse gas emissions for high-methane-emitting peatlands. In microcosms, catechin drastically reduced methane emissions by 72%-84% compared to controls. Longitudinal sampling allowed for reconstruction of a catechin degradation pathway involving Actinomycetota and Clostridium, which break down catechin into smaller phenolic compounds within the first 21 days, followed by degradation of phenolic compounds by Pseudomonas_E from Days 21 to 35. These genomes co-expressed hydrogen-uptake genes, suggesting hydrogenases may act as a hydrogen sink during catechin degradation and consequently reduce hydrogen availability to methanogens. In support of this idea, there was decreased gene expression by hydrogenotrophic and hydrogen-dependent methylotrophic methanogens under catechin treatment. There was also reduced gene expression from genomes inferred to be functioning syntrophically with hydrogen-utilizing methanogens. We propose that catechin metabolic redirection effectively starves hydrogen-utilizing methanogens, offering a potent avenue for curbing methane emissions across diverse environments including ruminants, landfills, and constructed or managed wetlands.
Keywords: ecology; metabolism; metagenome; metatranscriptome; methane; methanogens; microbiomes; polyphenols; rumen; wetlands.
© The Author(s) 2025. Published by Oxford University Press on behalf of the International Society for Microbial Ecology.
Conflict of interest statement
The authors declare no competing financial interests.
Figures


Update of
-
Polyphenol rewiring of the microbiome reduces methane emissions.bioRxiv [Preprint]. 2024 Oct 22:2024.10.22.619724. doi: 10.1101/2024.10.22.619724. bioRxiv. 2024. Update in: ISME J. 2025 Jan 2;19(1):wraf108. doi: 10.1093/ismejo/wraf108. PMID: 39484614 Free PMC article. Updated. Preprint.
References
-
- Myhre G, Shindell D, Bréon F et al. Anthropogenic and natural radiative forcing. In: Stocker TF, Qin D, Plattner GK et al (eds.), Climate change 2013: The Physical Science Basis. Contribution of Working Group I to the Fifth Assessment Report of the Intergovernmental Panel on Climate Change. Cambridge: Cambridge University Press, 2013, 659–740.
-
- Nzotungicimpaye C-M, MacIsaac AJ, Zickfeld K. Delaying methane mitigation increases the risk of breaching the 2°C warming limit. Commun Earth Environ 2023;4:250. 10.1038/s43247-023-00898-z - DOI
-
- European Commission. Launch by United States, the European Union, and Partners of the Global Methane Pledge to Keep 1.5C Within Reach. https://ec.europa.eu/commission/presscorner/detail/en/STATEMENT_21_5766 (22 October 2024, date last accessed).
-
- Saunois M, Stavert AR, Poulter B et al. The global methane budget 2000–2017. Earth Syst Sci Data 2020;12:1561–623. 10.5194/essd-12-1561-2020 - DOI
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

