An experimental test of the influence of microbial manipulation on sugar kelp (Saccharina latissima) supports the core influences host function hypothesis
- PMID: 40439420
- PMCID: PMC12175540
- DOI: 10.1128/aem.00301-25
An experimental test of the influence of microbial manipulation on sugar kelp (Saccharina latissima) supports the core influences host function hypothesis
Abstract
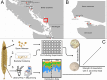
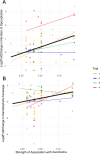
Kelp are valued for a wide range of commercial products and their role in kelp forest ecosystems, making kelp cultivation a rapidly expanding economic sector. Microbes associated with kelp and other macroalgae play a critical role in processes such as nutrient exchange, chemical signaling, and defense against pathogens. Thus, manipulating the microbiome to enhance macroalgal growth and resilience is a promising yet underexplored approach for sustainable kelp cultivation. The core microbiome hypothesis suggests that the bacteria that are consistently found on a host (the core microbes) are likely to have a disproportionate impact on host biology, making them an attractive target for microbiome manipulation. In this study, we surveyed wild Saccharina latissima and their surrounding environment to identify core bacterial taxa, compared them to cultivated kelp, and experimentally tested how cultured bacterial isolates affect kelp development. We found that core bacteria are nearly absent in cultivated juvenile sporophytes in nurseries, but eventually colonize them after outplanting to ocean farm sites. Bacterial inoculants had both positive and negative effects on kelp development. Notably, the strength of association of a bacterial genus with kelp in the wild positively correlated with its impact on gametophyte settlement and sporophyte development in kelp co-culture experiments, aligning with predictions from the core microbiome influences host function hypothesis. These findings affirm the feasibility of using microbial manipulations to improve current kelp aquaculture practices and provide a framework for developing these techniques.
Importance: Microorganisms consistently associated with hosts are widely thought to be more likely to impact host biology and health. However, this intuitive concept has not been experimentally evaluated. This study formalizes this concept as the Core Microbiome Influences Host Function hypothesis and experimentally tests this hypothesis in sugar kelp (Saccharina). The distribution of bacteria on wild kelp and core microbes was first identified by compiling a broad dataset of the kelp microbiome sampled across space and time. Bacterial cultures were isolated from the surface of sugar kelp and individually grown in laboratory co-culture with sugar kelp spores to assess the ability of bacterial isolates to influence kelp growth and development. In support of the core influences host function hypothesis, isolates belonging to bacterial genera that are more strongly associated with wild sugar kelp are more likely to influence development in laboratory experiments.
Keywords: Saccharina latissima; bacterial isolation; core microbiome; kelp cultivation; microbial ecology; microbial manipulation.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





Similar articles
-
Abiotic stress destabilizes the bacterial community of sugar kelp, Saccharina latissima (Phaeophyceae).J Phycol. 2025 Aug;61(4):840-857. doi: 10.1111/jpy.70033. Epub 2025 May 28. J Phycol. 2025. PMID: 40432552 Free PMC article.
-
Ocean acidification significantly alters the trace element content of the kelp, Saccharina latissima.Mar Pollut Bull. 2024 May;202:116289. doi: 10.1016/j.marpolbul.2024.116289. Epub 2024 Apr 1. Mar Pollut Bull. 2024. PMID: 38564822
-
A bloom of a single bacterium shapes the microbiome during outdoor diatom cultivation collapse.mSystems. 2025 Jun 17;10(6):e0037525. doi: 10.1128/msystems.00375-25. Epub 2025 May 14. mSystems. 2025. PMID: 40366134 Free PMC article.
-
Assessing the comparative effects of interventions in COPD: a tutorial on network meta-analysis for clinicians.Respir Res. 2024 Dec 21;25(1):438. doi: 10.1186/s12931-024-03056-x. Respir Res. 2024. PMID: 39709425 Free PMC article. Review.
-
Symbiotic symphony: Understanding host-microbiota dialogues in a spatial context.Semin Cell Dev Biol. 2024 Sep-Oct;161-162:22-30. doi: 10.1016/j.semcdb.2024.03.001. Epub 2024 Apr 1. Semin Cell Dev Biol. 2024. PMID: 38564842 Review.
References
-
- Adams JM, Gallagher JA, Donnison IS. 2009. Fermentation study on Saccharina latissima for bioethanol production considering variable pre-treatments. J Appl Phycol 21:569–574. doi: 10.1007/s10811-008-9384-7 - DOI
-
- Marinho GS, Holdt SL, Birkeland MJ, Angelidaki I. 2015. Commercial cultivation and bioremediation potential of sugar kelp, Saccharina latissima, in Danish waters. J Appl Phycol 27:1963–1973. doi: 10.1007/s10811-014-0519-8 - DOI
-
- Wargacki AJ, Leonard E, Win MN, Regitsky DD, Santos CNS, Kim PB, Cooper SR, Raisner RM, Herman A, Sivitz AB, Lakshmanaswamy A, Kashiyama Y, Baker D, Yoshikuni Y. 2012. An engineered microbial platform for direct biofuel production from brown macroalgae. Science 335:308–313. doi: 10.1126/science.1214547 - DOI - PubMed
-
- Bak UG, Nielsen CW, Marinho GS, Gregersen Ó, Jónsdóttir R, Holdt SL. 2019. The seasonal variation in nitrogen, amino acid, protein and nitrogen-to-protein conversion factors of commercially cultivated Faroese Saccharina latissima. Algal Res 42:101576. doi: 10.1016/j.algal.2019.101576 - DOI
-
- Forbord S, Steinhovden KB, Solvang T, Handå A, Skjermo J. 2020. Effect of seeding methods and hatchery periods on sea cultivation of Saccharina latissima (Phaeophyceae): a norwegian case study. J Appl Phycol 32:2201–2212. doi: 10.1007/s10811-019-01936-0 - DOI
MeSH terms
Supplementary concepts
LinkOut - more resources
Full Text Sources

