Application of a high-resolution melt assay for monitoring SARS-CoV-2 variants in Burkina Faso and Kenya
- PMID: 40439429
- PMCID: PMC12188703
- DOI: 10.1128/msphere.00027-25
Application of a high-resolution melt assay for monitoring SARS-CoV-2 variants in Burkina Faso and Kenya
Abstract
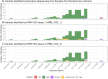
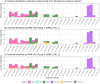
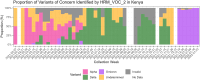
The rapid emergence and global dissemination of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) highlighted a need for robust, adaptable surveillance systems. However, financial and infrastructure requirements for whole-genome sequencing mean most surveillance data have come from higher-resource geographies, despite unprecedented investment in sequencing in low- and middle-income countries (LMICs). Consequently, the molecular epidemiology of SARS-CoV-2 in some LMICs is limited, and there is a need for more cost-accessible technologies to help close data gaps for surveillance of SARS-CoV-2 variants. To address this, we have developed two high-resolution melt (HRM) curve assays that target variant-defining mutations in the SARS-CoV-2 genome, which give unique signature profiles that define different SARS-CoV-2 variants of concern (VOCs). Extracted RNA from SARS-CoV-2-positive samples collected from 205 participants (112 in Burkina Faso, 93 in Kenya) enrolled in the MALCOV study (Malaria as a Risk Factor for COVID-19) between February 2021 and February 2022 were analyzed using our optimized HRM assays. With next-generation sequencing on Oxford Nanopore MinION as a reference, two HRM assays, HRM-VOC-1 and HRM-VOC-2, demonstrated sensitivity/specificity of 100%/99.29% and 92.86%/99.39%, respectively, for detecting Alpha, 90.08%/100% and 92.31%/100% for Delta, and 93.75%/100% and 100%/99.38% for Omicron BA.1. The assays described here provide a lower-cost approach to conducting molecular epidemiology, capable of high-throughput testing. We successfully scaled up the HRM-VOC-2 assay to screen a total of 506 samples from which we were able to show the replacement of Alpha with the introduction of Delta and the replacement of Delta by the Omicron variant in this community in Kisumu, Kenya.IMPORTANCEThe rapid evolution of the severe acute respiratory syndrome coronavirus 2 variants of concern (VOCs) demonstrated the need for accessible surveillance tools so all communities can conduct viral surveillance. Sequencing, the gold standard, is still a largely inaccessible methodology in low-resource settings. Here, we present a quick, low-cost tool to screen for the common VOCs, designed to support surveillance efforts in low-resource settings. This tool was used to screen samples from Burkina Faso and Western Kenya throughout the pandemic. We show through comparison to sequencing that our assay can generate highly similar data on the different variants circulating in a population, therefore showing the effectiveness of our tool. While not a replacement for sequencing, we present a method of screening and prioritizing samples for further investigation and reduce overburdening sequencing capacity. Our findings provide insight into one potential tool that could be further applied to pathogen screening in the absence of robust sequencing infrastructure.
Keywords: Africa; Burkina Faso; COVID-19; HRM; Kenya; SARS-CoV-2; diagnostics; surveillance; variants of concern.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
Rapid, point-of-care antigen tests for diagnosis of SARS-CoV-2 infection.Cochrane Database Syst Rev. 2022 Jul 22;7(7):CD013705. doi: 10.1002/14651858.CD013705.pub3. Cochrane Database Syst Rev. 2022. PMID: 35866452 Free PMC article.
-
Antibody tests for identification of current and past infection with SARS-CoV-2.Cochrane Database Syst Rev. 2022 Nov 17;11(11):CD013652. doi: 10.1002/14651858.CD013652.pub2. Cochrane Database Syst Rev. 2022. PMID: 36394900 Free PMC article.
-
Signs and symptoms to determine if a patient presenting in primary care or hospital outpatient settings has COVID-19.Cochrane Database Syst Rev. 2022 May 20;5(5):CD013665. doi: 10.1002/14651858.CD013665.pub3. Cochrane Database Syst Rev. 2022. PMID: 35593186 Free PMC article.
-
Measures implemented in the school setting to contain the COVID-19 pandemic.Cochrane Database Syst Rev. 2022 Jan 17;1(1):CD015029. doi: 10.1002/14651858.CD015029. Cochrane Database Syst Rev. 2022. Update in: Cochrane Database Syst Rev. 2024 May 2;5:CD015029. doi: 10.1002/14651858.CD015029.pub2. PMID: 35037252 Free PMC article. Updated.
-
Nanopore sequencing reveals the genomic diversity of the variants of concern of SARS-CoV-2 during 2021 disease outbreak in Pakistan.Sci Rep. 2025 Aug 1;15(1):28129. doi: 10.1038/s41598-025-12774-1. Sci Rep. 2025. PMID: 40750817 Free PMC article.
References
-
- World Health Organization . Tracking SARS-CoV-2 variants. Accessed 9 June 2023. https://www.who.int/activities/tracking-SARS-CoV-2-variants.
-
- Genomic sequencing effort for SARS-CoV-2 by country during the pandemic. Elsevier Enhanced Reader [Internet]. Available from: https://reader.elsevier.com/reader/sd/pii/S1201971220325571?token=E1DF89.... Retrieved 2222 DecDecember 2022. Accessed , 2222 DecDecember 2022
-
- WHO, Regional Office for Africa [Internet]. 2023. Scaling up genomic sequencing in Africa. Available from: https://www.afro.who.int/news/scaling-genomic-sequencing-africa
MeSH terms
Substances
Supplementary concepts
Grants and funding
- INV-019400/GATES/Gates Foundation/United States
- INV-017985,INV-019400/Bill and Melinda Gates Foundation
- NIHR200907/National Institute for Health Research Health Protection Research Unit
- 75F40120C00085/U.S Food and Drug Administration Medical Countermeasures Initiative Contract
- INV-017985/GATES/Gates Foundation/United States
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous
