Computational Design of Lysine Targeting Covalent Binders Using Rosetta
- PMID: 40440538
- PMCID: PMC12152945
- DOI: 10.1021/acs.jcim.5c00212
Computational Design of Lysine Targeting Covalent Binders Using Rosetta
Abstract
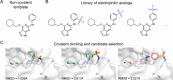
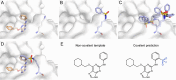
Chemical probes that form a covalent bond with their target protein have been established as a powerful tool for investigating proteins and modulating their activity, but until recently were mostly targeting cysteine residues. Covalent binders that target lysine residues are increasingly reported. Covalent binding to lysine involves challenges such as the increased pKa of the side chain and its considerable flexibility. Here, we describe two computational methods to derivatize lysine-binding covalent small-molecules based on known noncovalent binders, approaching the design problem from two opposite directions. In a "ligand-side" approach, we scan different ligand positions to install an electrophile and dock these derivatized ligands into the target protein. In a "protein-side" approach, we install an electrophile on the target lysine and model its conformational space to find suitable installation vectors on the ligand. We applied both of these protocols retrospectively to a data set of electrophilic ligands and to a data set of vitamin B6 covalently bound to a receptor lysine residue. Our ligand-side protocol successfully identified the known covalent binder in 80% and 86% of cases, while the protein-side protocol achieved identification rates of 56% and 82%, respectively. We prospectively validated these protocols by designing and testing a new lysine-targeting MKK7 inhibitor. Mass-spectrometry and crystallography validated the covalent binding to the target lysine. Applying these protocols to a data set of known kinase inhibitors identified high-confidence covalent candidates for more than 200 human kinases, demonstrating the potential impact of our protocols.
Figures


Similar articles
-
Covalent flexible peptide docking in Rosetta.Chem Sci. 2021 Jul 12;12(32):10836-10847. doi: 10.1039/d1sc02322e. eCollection 2021 Aug 18. Chem Sci. 2021. PMID: 34476063 Free PMC article.
-
A simple method for developing lysine targeted covalent protein reagents.Nat Commun. 2023 Dec 1;14(1):7933. doi: 10.1038/s41467-023-42632-5. Nat Commun. 2023. PMID: 38040731 Free PMC article.
-
Lysine-Targeted Inhibitors and Chemoproteomic Probes.Annu Rev Biochem. 2019 Jun 20;88:365-381. doi: 10.1146/annurev-biochem-061516-044805. Epub 2019 Jan 11. Annu Rev Biochem. 2019. PMID: 30633551 Review.
-
Inhibition of Mcl-1 through covalent modification of a noncatalytic lysine side chain.Nat Chem Biol. 2016 Nov;12(11):931-936. doi: 10.1038/nchembio.2174. Epub 2016 Sep 5. Nat Chem Biol. 2016. PMID: 27595327
-
Arylfluorosulfate-Based Electrophiles for Covalent Protein Labeling: A New Addition to the Arsenal.Angew Chem Int Ed Engl. 2019 Jan 21;58(4):957-966. doi: 10.1002/anie.201806037. Epub 2018 Oct 26. Angew Chem Int Ed Engl. 2019. PMID: 30024079 Free PMC article. Review.
References
-
- Bradshaw J. M., McFarland J. M., Paavilainen V. O., Bisconte A., Tam D., Phan V. T., Romanov S., Finkle D., Shu J., Patel V., Ton T., Li X., Loughhead D. G., Nunn P. A., Karr D. E., Gerritsen M. E., Funk J. O., Owens T. D., Verner E., Brameld K. A., Hill R. J., Goldstein D. M., Taunton J.. Prolonged and Tunable Residence Time Using Reversible Covalent Kinase Inhibitors. Nat. Chem. Biol. 2015;11(7):525–531. doi: 10.1038/nchembio.1817. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

