Lipids modulate the dynamics of GPCR:β-arrestin interaction
- PMID: 40442125
- PMCID: PMC12122939
- DOI: 10.1038/s41467-025-59842-8
Lipids modulate the dynamics of GPCR:β-arrestin interaction
Abstract
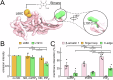
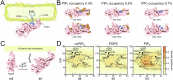
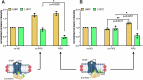
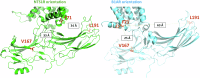
β-arrestins are key molecular partners of G Protein-Coupled Receptors (GPCRs), triggering not only their desensitization but also intracellular signaling. Existing structural data point to high conformational plasticity of GPCR:β-arrestin interaction, with two completely different orientations between receptor and β-arrestin. Combining molecular dynamics simulations and fluorescence spectroscopy, we show that β-arrestin 1 interacts with membranes even in the absence of a receptor, an interaction that is enhanced by PI(4,5)P2, presumably holding the β-arrestin 1 C-edge loop into the lipid bilayer. This key interaction helps β-arrestin 1 to adopt a "receptor-ready" orientation and consequently favors its coupling to the ghrelin receptor (GHSR). In addition, we show that the GHSR:β-arrestin 1 assembly is a dynamic complex where β-arrestin can adopt several orientations. PI(4,5)P2 decreases the dynamics of the complex and shifts the equilibrium between the different arrangements, favoring one of them. Taken together, our results highlight how PI(4,5)P2 plays a true third-player role in the GPCR:β-arrestin interaction, not only by preparing β-arrestin for its further interaction with receptors but also by modulating its orientation once the protein:protein complex is formed.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures


References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

