Early mucosal responses following a randomised controlled human inhaled infection with attenuated Mycobacterium bovis BCG
- PMID: 40442144
- PMCID: PMC12122720
- DOI: 10.1038/s41467-025-60285-4
Early mucosal responses following a randomised controlled human inhaled infection with attenuated Mycobacterium bovis BCG
Abstract
The development of an effective vaccine against Mycobacterium tuberculosis is hampered by an incomplete understanding of immunoprotective mechanisms. We utilise an aerosol human challenge model using attenuated Mycobacterium bovis BCG, in BCG-naïve UK adults. The primary endpoint of this study (NCT03912207) was to characterise the early immune responses induced by aerosol BCG infection, the secondary endpoint was to identify immune markers associated with in-vitro protection. Blinded volunteers were randomised to inhale 1 × 107 CFU aerosolised BCG or 0.9% saline (20:6); and sequentially allocated to bronchoscopy at day 2 or 7 post-inhalation (10 BCG, 3 saline each timepoint). In the bronchoalveolar lavage post-aerosol BCG infection, there was an increase in frequency of eosinophils, neutrophils, NK cells and Donor-Unrestricted T cells at day 7, and the frequency of antigen presenting cells decreased at day 7 compared with day 2. The frequency of interferon-gamma+ BCG-specific CD4+ T cells increased in the BAL and peaked in the blood at day 7 post-BCG infection compared to day 2. BAL cells at day 2 and day 7 upregulated gene pathways related to phagocytosis, MHC-II antigen loading, T cell activation and proliferation. BCG's lack of key virulence factors and its failure to induce granulomas, may mean the observed immune responses do not fully recapitulate Mycobacterium tuberculosis infection. However, human infection models can provide unique insights into early immune mechanisms, informing vaccine design for complex pathogens.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures



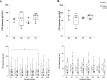

References
-
- Schrager, L. K. et al. WHO preferred product characteristics for new vaccines against tuberculosis. Lancet Infect. Dis.18, 828–829 (2018). - PubMed
-
- Fletcher, H. A. & Dockrell, H. M. Human biomarkers: can they help us to develop a new tuberculosis vaccine?. Future Microbiol11, 781–787 (2016). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

